Rare cancers account for about 25% of cancer in adults and about 10% of pediatric cancer cases. Unfortunately, prognosis for rare types of cancer is generally worse than for common cancers, not necessarily because these types of rare cancer are more aggressive, but because there is not a lot of preclinical or clinical data that can guide a doctor’s treatment plan. Without empirical data to drive their decisions, doctors are forced to rely on anecdotal evidence and clinical intuition to treat these patients. Obviously, this is not an ideal situation. A large collaborative effort between research groups at Boston Children’s Hospital, Dana-Farber Cancer Institute, the Broad Institute, the Koch Institute, and Brigham and Women’s Hospital in Boston recently published a proof-of-concept study to find druggable targets in rare cancers.
It is now easier than ever to create a patient-derived cell line, giving researchers highly specific and personalized tool to figure out treatments on cells from specific tumors. This proof-of-concept study published in
Nature Communications created and validated a patient-derived cell line from a pediatric patient with a rare form of cancer and then used CRISPR-Cas9, RNA interference, and pharmacological screens to identify novel, druggable targets to guide treatment.
The first step was to create a propagating cell line from a pediatric patient with a rare undifferentiated sarcoma. By comparing their cell line to biopsy tissue from the patient, the researchers proved that their cell line faithfully recapitulated the histology, immunohistochemistry, and somatic mutations of the tumor. Additionally, when the patient-derived cells were injected into immunocompromised mice, they formed tumors at similar rates to established cancer cell lines.
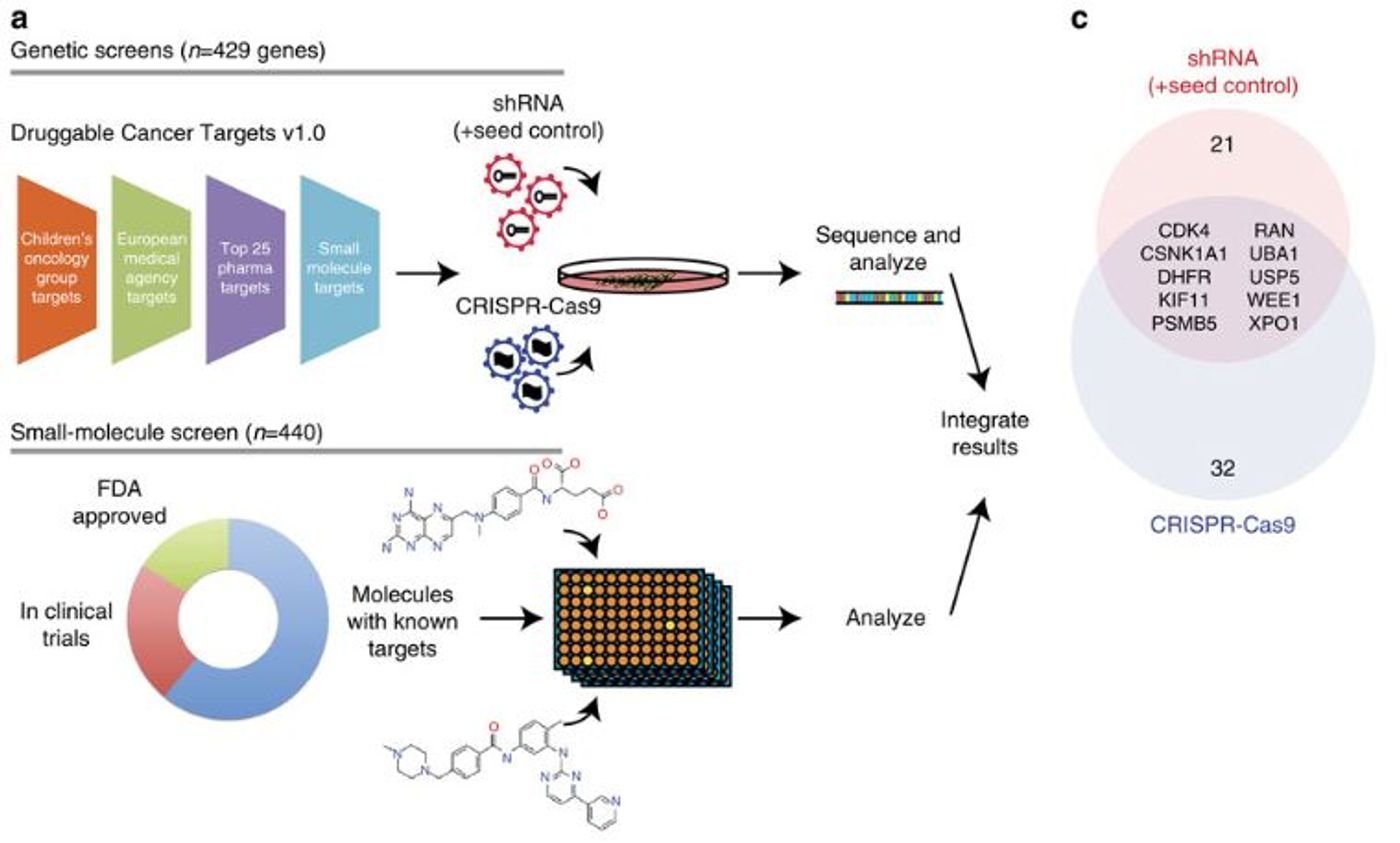
Once the cell line was thoroughly vetted, the researchers used CRISPR-Cas9 genome editing and RNA interference to identify “dependencies” in the cancer cells. In this context, dependencies refer to genes that when deleted, suppressed, or inhibited lead to decreased proliferation or survival. CRISPR-Cas9 and RNA interference were both used because they have different cellular effects. With CRISPR-Cas9, the gene is completely deleted, but with RNA interference, the gene still exists and is transcribed into mRNA, but is not processed or translated into a functional protein. Both approaches were used in this study because dependencies that were found in both screens would be the most robust targets.
Between both of the screens, the researchers identified ten genes that reduced cancer cell survival when deleted or suppressed. These ten genes were then further tested with pharmacological screening with drugs that are either approved, up for approval, or are in the pharmaceutical pipeline. Adding the drug screening to the gene suppression data, the researchers pulled out two targets, CDK4 and XPO1, that can be targeted with small molecules or gene suppression to reduce the proliferation and survival of these specific cancer cells.
What is interesting is that neither of these two genes are genetically associated with this type of tumor and would not have been identified by just sequencing the genome. The gene suppression studies combined with the pharmacological screen was able to pull out these two novel, druggable targets that would have otherwise been missed.
This proof-of-concept study was a massive undertaking and required an insane amount of work, but it does provide a workflow for identifying therapeutic targets in rare, under-researched cancers. Creating a cell line from a patient’s unique cancer, identifying which genes are required for those cells to survive, and screening for available drugs to target those genes is the epitome of personalized medicine.