A Critical Step in Ribosome Assembly is Viewed For the First Time
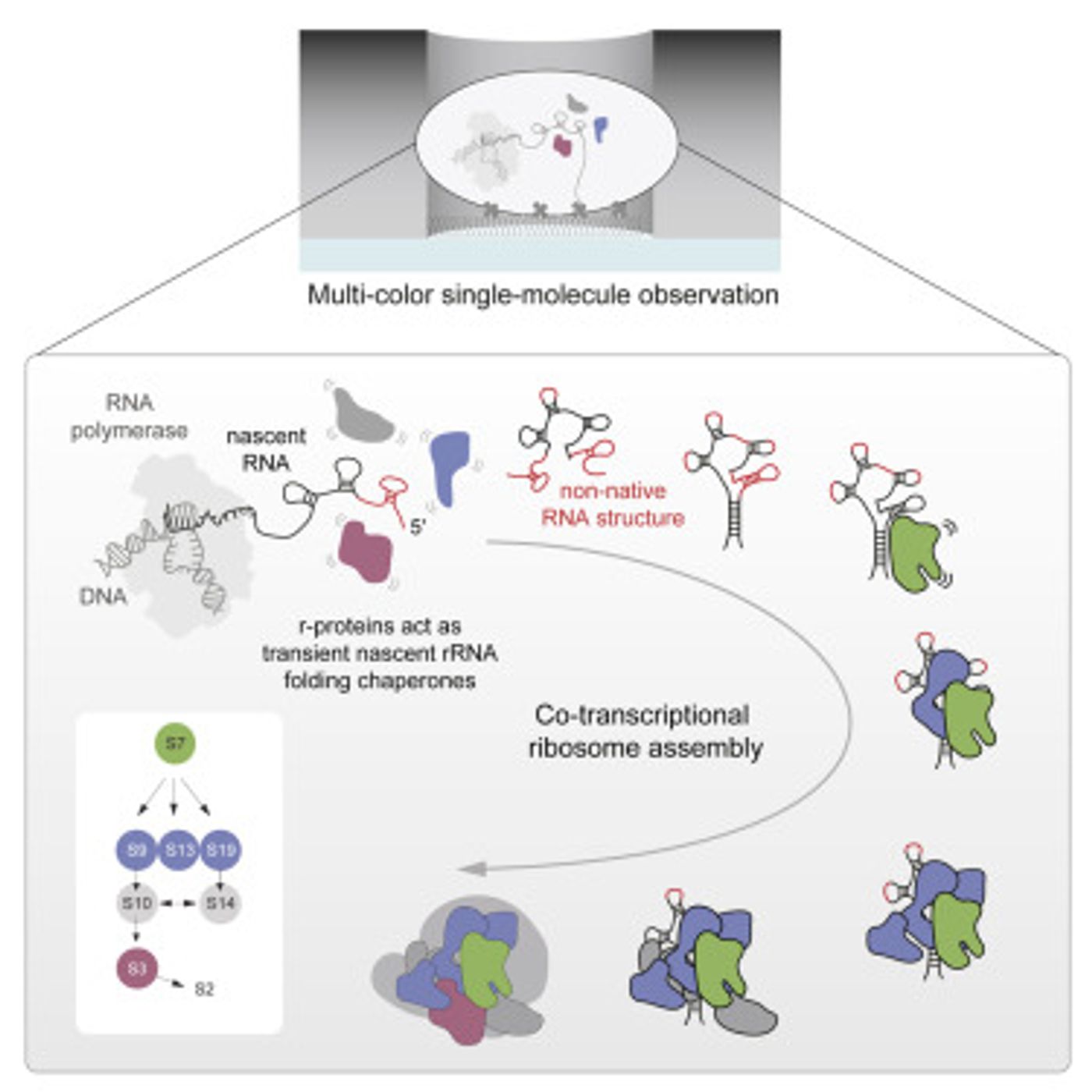
Proteins are critical to biology, carrying out many functions and giving rise to crucial structures. These essential molecules are generated by vital, ancient cellular structures called ribosomes (which are described in the video). In a first, a research team from Scripps Research Institute and Stanford University has captured in real-time an important part of the assembly of ribosomes. The findings, which have shown how strands of genetic material called RNA are coaxed into properly folding to form a component of ribosomes, have been reported in Cell. This work also shows that ribosomes are not, as was thought, generated in a series of steps.
"In contrast to what had been the dominant theory in the field, we revealed a far more chaotic process," noted James R. Williamson, Ph.D., a professor in the Department of Integrative Structural & Computational Biology at Scripps Research. "It's not a sleek Detroit assembly line--it's more like a trading pit on Wall Street."
In this study, the scientists used a technique called zero-mode waveguide single-molecule fluorescence microscopy to track RNAs and proteins in real-time.
"This shows that we now can examine in detail how RNAs fold while they are being synthesized and proteins are assembling on them," said the first author of the study Olivier Duss, Ph.D., a postdoctoral research fellow in the Department of Integrative Structural & Computational Biology at Scripps Research. "This has been a very difficult thing to study in biology because it involves several distinct biological processes that are dependent on each other and have to be detected simultaneously."
The research builds on the team's previous work that recorded a well-understood step in ribosome assembly. Now they have gone further and revealed more about RNA folding and the guidance it gets from eight proteins that become part of the E. Coli ribosome. The proteins interact temporarily with the RNA multiple times before eventually settling into their resting places in the RNA-protein complex.
"Our study indicates that in ribosomal RNA-folding, and perhaps more generally in RNA-folding in cells, many proteins help fold RNA though weak, transient and semi-specific interactions with it," Duss noted.
The work, which reveals more about a very basic process, may have broader implications for medicine. Some antibiotics work by disrupting ribosomes in bacteria, and future antibiotics might target them more specifically, and cause fewer side effects. The research also establishes an approach for studying RNA molecules, many of which can be present in a cell at any one time. It can also provide insight into how RNA can misfold, or how that dysfunction can be corrected. Many diseases are thought to be related to aberrantly folded RNA.
"When we understand more fully how bacterial ribosomes assemble and function, we could potentially target them in ways that affect a narrower group of harmful bacterial species and spare the good ones, reducing side effects for patients," Duss explained. Some cancer drugs slow the formation of ribosomes as well. If we know more about ribosome assembly in humans, we may be able to stop the growth of cancer more precisely and efficiently, Duss added.
The team now wants to learn more about the remaining steps in ribosome production, and other kinds of interactions between proteins and RNA.
Sources: AAAS/Eurekalert! via Scripps Research Institute, Cell