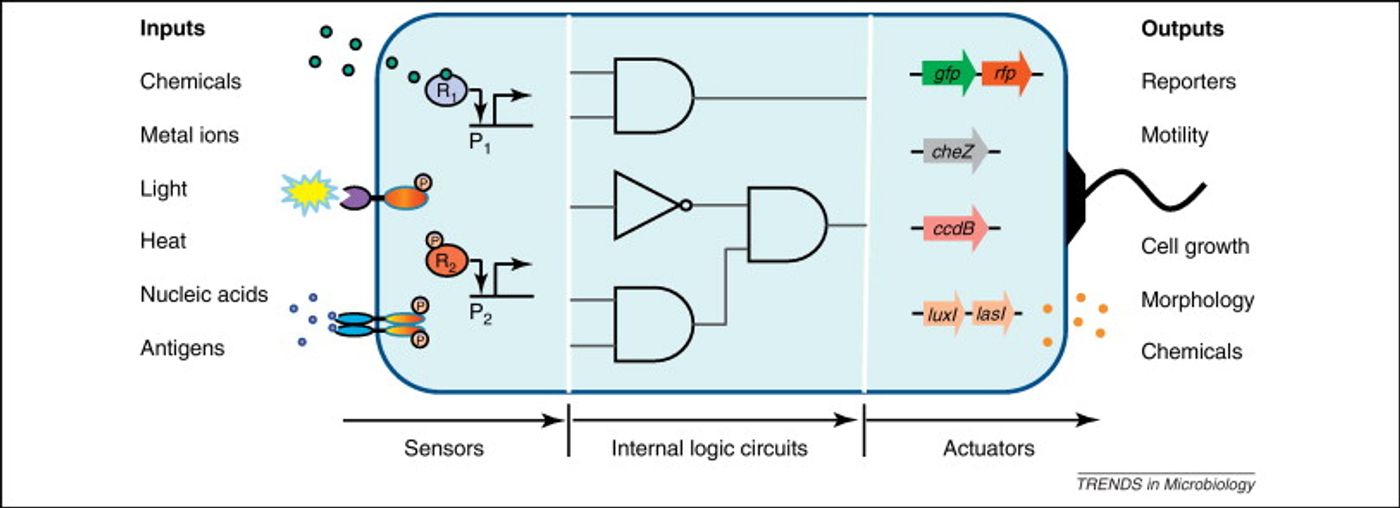
Cells in nature have to respond to environmental stimuli with a complex repertoire of responses. In effect, they make computations that require processing that is both analog, like adjusting to a gradual change in temperature, or digital in nature, like when a cell initiates division or death.
In contrast, as synthetic systems modeling biology have been developed, the focus has been on replicating either digital or analogue processing, not both at once. This has made for limitations in their use. Innovative work done by a group at Massachusetts Institute of Technology (MIT) and
published in Nature Communications now demonstrates the integration of those two disparate types of actions. That breakthrough will allow for the creation of gene circuits that are adept at performing complicated computing tasks.
Such synthetic circuitry would thus be able to sense the magnitude of some analogue input, make a determination about the proper reaction to that level, and then have an appropriate output to the situation.
For example, circuits that might be designed in the future may be used to monitor levels of glucose in the blood, and then respond in a variety of ways depending on what the concentration was. “If the glucose level was too high you might want your cells to produce insulin, if the glucose was too low you might want them to make glucagon, and if it was in the middle you wouldn’t want them to do anything,” says Timothy Lu, Associate Professor in the Department of Electrical Engineering and Computer Science at MIT and leader of the research team that did this work.
“Most of the work in synthetic biology has focused on the digital approach, because [digital systems] are much easier to program,” Lu says.
Digital computing systems are based on binary output like 0 or 1, so executing complex operations requires a large number of parts to carry them out, a feature that is very challenging for artificial biological systems.
“Digital is basically a way of computing in which you get intelligence out of very simple parts, because each part only does a very simple thing, but when you put them all together you get something that is very smart,” Lu explains. “But that requires you to be able to put many of these parts together, and the challenge in biology, at least currently, is that you can’t assemble billions of transistors like you can on a piece of silicon,” he continues.
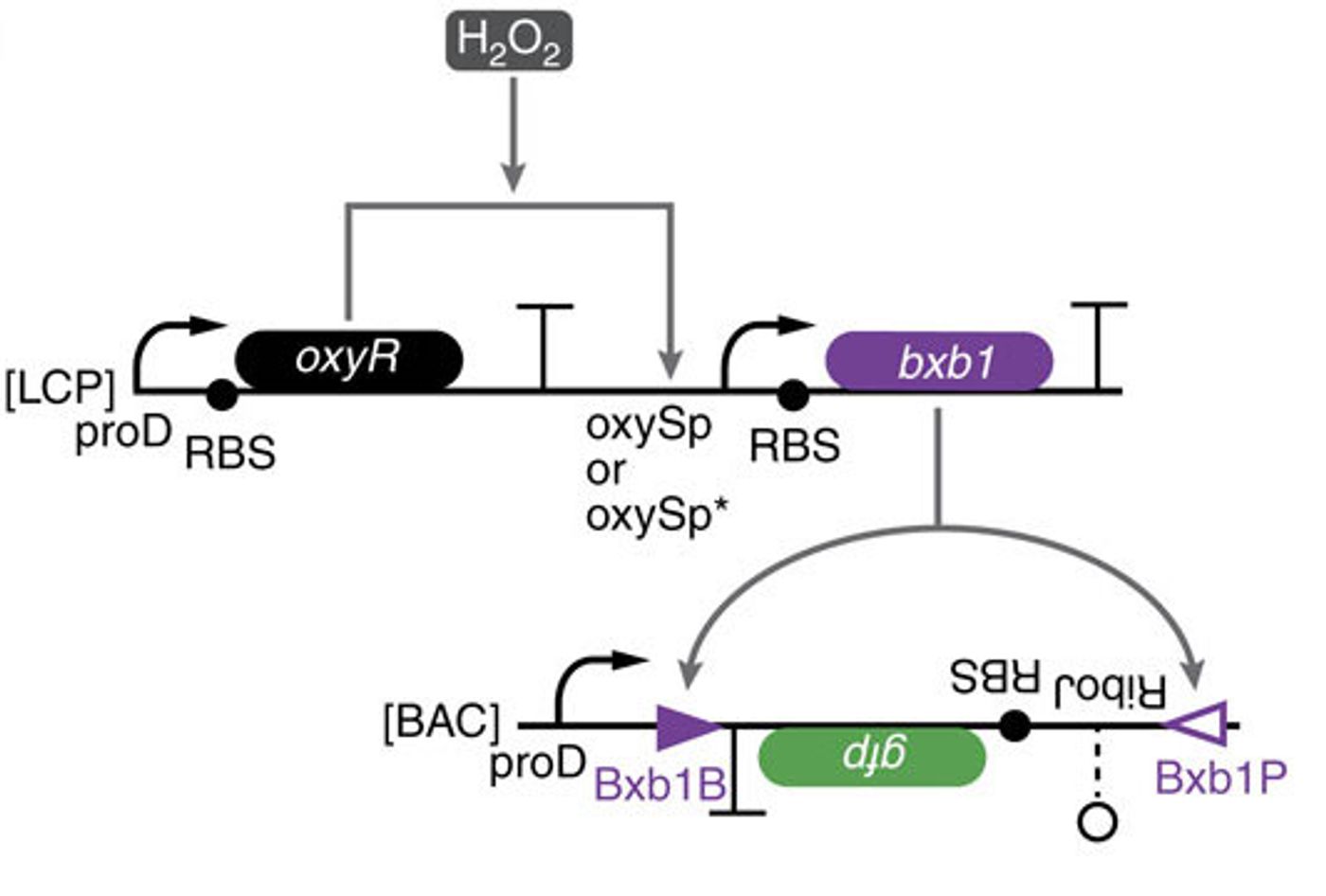
The scientists thus created a module with multiple elements. One will sense amounts – an analogue input – and the other element controls the response – the digital output. In this case, the response element is a recombinase gene, capable of turning expression of a particular gene on or off by inverting it. So to put it all together, the concentration of a chemical reaches a particular level, causing the threshold module to express the recombinase, which flips the DNA segment. That segment contains a regulatory element or gene that then changes the expression of some specific output.
“So this is how we take an analogue input, such as a concentration of a chemical, and convert it into a 0 or 1 signal,” Lu says. “And once that is done, and you have a piece of DNA that can be flipped upside down, then you can put together any of those pieces of DNA to perform digital computing,” he explains.
For starters, the research team is testing the concept of using analogue-to-digital converters to monitor inflammation in the gut caused by dysfunction such as inflammatory bowel disease, and releasing varying amounts of a drug treatment in response.
Another idea is to engineer immune cells used for cancer treatment to detect different environmental conditions, like tumor lysis or oxygen levels, and alter their therapeutic action in response.
Learn more about engineering and programming gene circuits in the video below.
Sources:
MIT,
Nature Communications