A new sequencing technique has demonstrated that variations in RNA processing can reveal details about how development is regulated.
Organic life on earth depends on the translation of a genetic blueprint into proteins that carry out specific functions. Through the use of cellular machinery, DNA is transcribed into messenger RNA (mRNA) that encodes for a protein, made up of amino acids and using transfer RNAs (tRNAs). Poly(A) tails are added to the ends of mRNAs, in one of the processing steps following its transcription. They’re long chains of adenine nucleotides that increase the stability of the RNA to prevent degradation and seem to have a larger regulatory function. The video below from Khan Academy explains some details of post-transcriptional processing of RNA.
A new sequencing technique
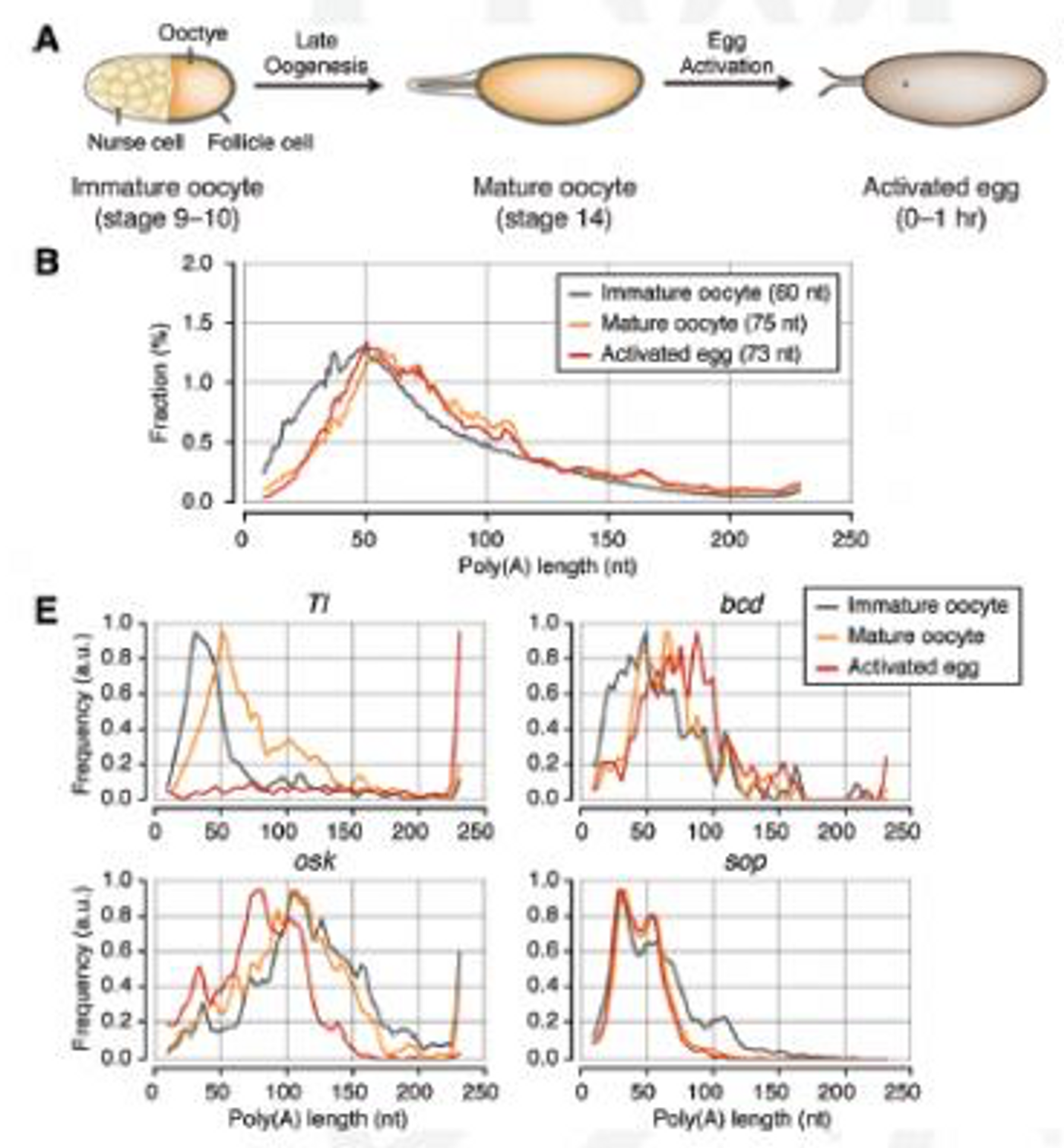
described in a publication in Genes and Development by Seoul’s Institute for Basic Science Center for RNA Research focuses on the investigation of poly(A) tails in gene regulation. The research team focused on Drosophila embryos and oocytes to get a view of the developmental landscape.
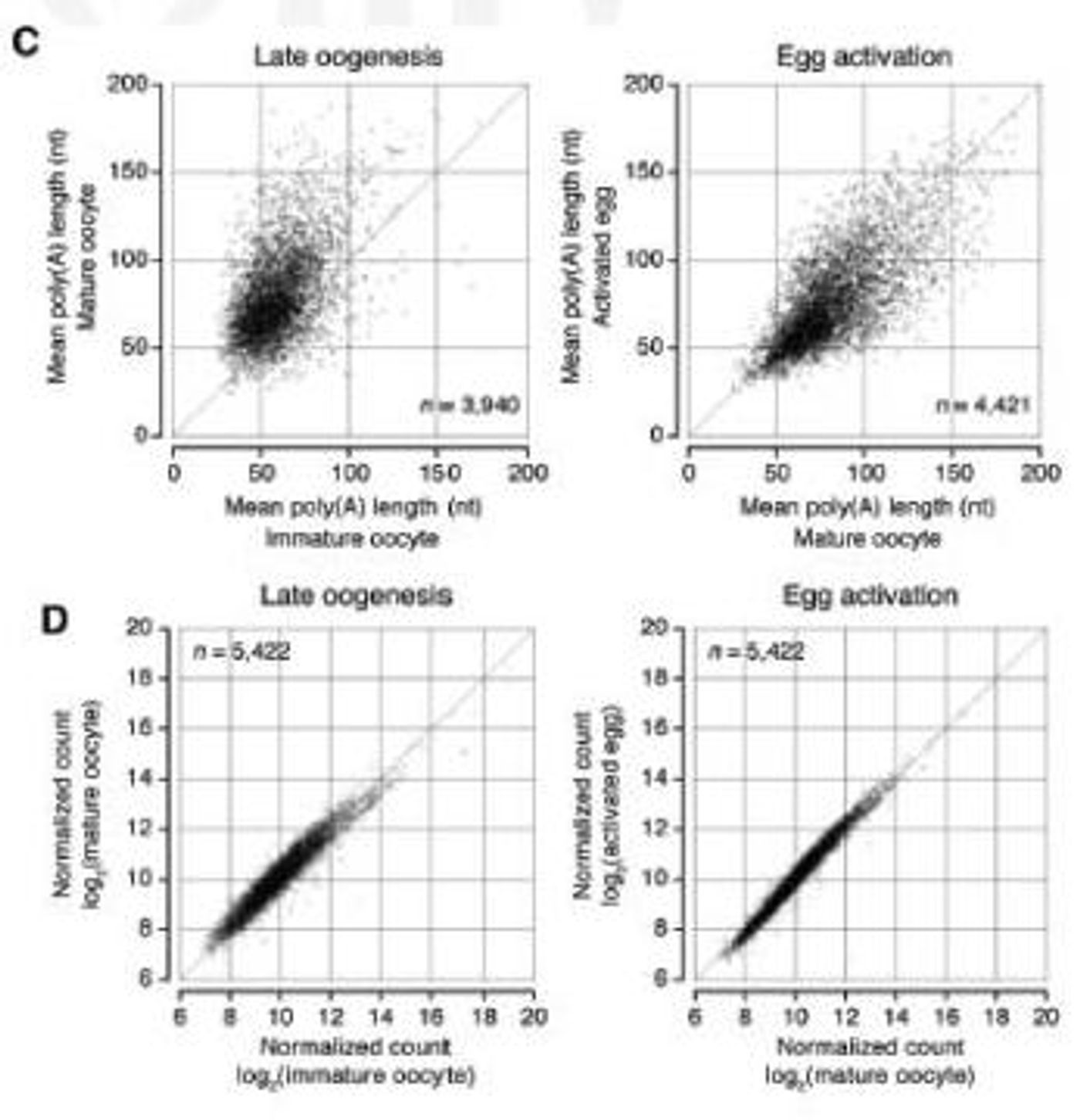
In oocytes, mRNAs lie dormant, waiting for activation and translation into proteins. There, long poly(A) tails are added to the mRNAs that are necessary for the early stages of development; the team wanted to see exactly how the length of these tails influenced gene expression. The scientists had previously created TAIL-seq for that purpose, and now have vastly improved its sensitivity with a new version they call mTAIL-seq, which has a sequencing depth enhancement of about 1000-fold.
First author Jaechul Lim explains, "We used mTAIL-seq to measure poly(A) length of maternal mRNAs in oocyte-to-embryo development. From the genomic scale analysis, we found global dynamic poly(A) tail regulation without the change of mRNA abundance."
Another way to analyze translation is to check the efficiency of translation of mRNA into protein - ribosomal profiling (RPF). The investigators compared RPF data to the mTAIL-seq data and found a strong correlation between poly(A) tail length and translational efficiency in the early embryo. The researchers suggest that could indicate poly(A) tails thus aid in the direction of animal development.
"The global profiling of poly(A) tails by mTAIL-seq provides a comprehensive resource for the regulation of poly(A) tails in Drosophila oocyte-to-embryo development and it help us to understand how poly(A) tail of maternal mRNA affect the production of proteins at the beginning of embryonic development," said another author of the study, Mihye Lee.
mTAIL-seq is described as a technique that boasts high sensitivity and low cost, alongside technical robustness and broad accessibility. As such the research team hopes that their invention will be used on a broad scale to improve the understanding of how mRNA tailing impacts a wide variety of biological systems.
Sources:
AAAS/Eurekaltert! via
IBS,
Genes and Development