Major advance demonstrated in X-ray crystallographic sample techniques
New research published in Nature Communications has corroborated a technique using a microfluidic droplet generator that is capable of shrinking sample size and waste in Serial Femtosecond Crystallography (SFX) experiments. The research was headed by scientists at Arizona State University’s School of Molecular Sciences (SMS), department of physics and the Biodesign Institute Center for Applied Structural Discovery.
Sample loss can reach as high as 99% in some X-ray crystallographic sample strategies, so improving those numbers is much to tout about. The development came about as a collaboration between ASU researchers and European researchers at the European X-ray Free Electron Laser (EuXFEL) research facility. Alexandra Ros, a professor in ASU's SMS, has been leading the project for years. She comments:
"We are excited that this work, resulting from a huge collaborative effort, has been well received in the XFEL community. We are further developing this method and are seeking synchronization of the microfluidic droplets with the pulses of XFELs. At this very moment, a small team of ASU students has just finished performing experiments at the Linac Coherent Light Source (LCLS) at the SLAC National Accelerator Laboratory in Menlo Park, CA to refine the method. There could not have been better timing for the publication of our work."
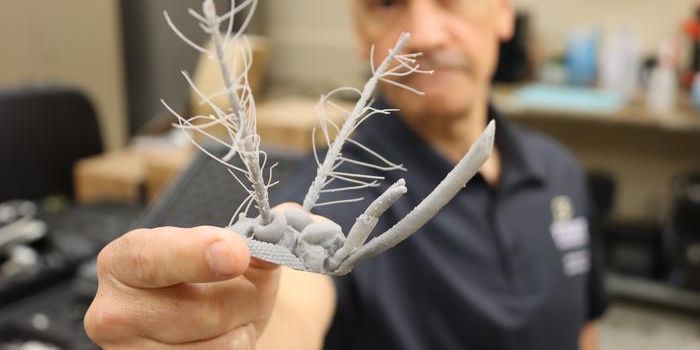
Serial femtosecond crystallography (SFX) combined with X-ray free electron lasers (XFELs) can determine the structures of membrane proteins and time-resolved crystallography. In SFX, reports Eureka Alert, a liquid stream with protein crystals is intersected with a high-intensity XFEL beam that is a billion times brighter than traditional synchrotron X-ray sources. The addition from Ros’s team of a high-resolution 3D-printed microfluidic device that generates aqueous-in-oil droplets synchronized to the free electron laser pulses increases the precision of SFX, therefore greatly reducing the need for large amounts of purified protein sample.
The method is particularly useful for membrane proteins and other hard-to-crystallize proteins because it provides high-resolution structural information from micro- and nanocrystals. In this most recent experiment, says the team, they were able to determine the crystal structure of the enzyme 3-Deoxy-d-manno-Octulosonate 8-Phosphate Synthase (KDO8PS), identifying a potential target for antibiotic studies. These results are so significant that they could change the way that scientists conduct SFX at XFELs around the world, consistently integrating droplet generation by segmented oil flow from here on out.
Sources: Nature Communications, Eureka Alert