Of Mice and Men: Deep Learning Transforms Diagnostics
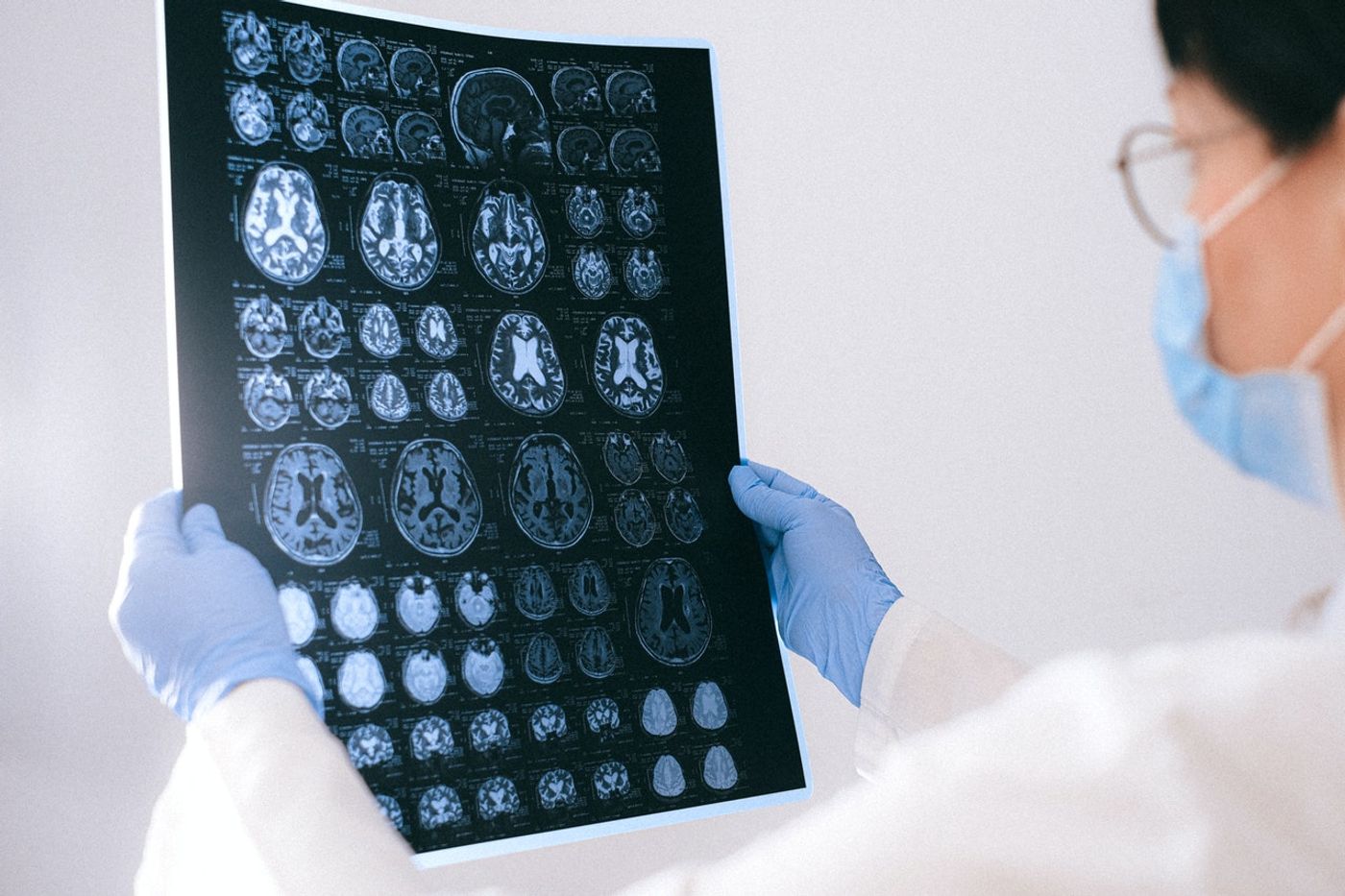
Medical imaging technologies enable physicians to take a peek under the hood, capturing snapshots of the internal organs and tissues to figure out what’s wrong. Inside their imaging toolboxes, clinicians have everything from “old school” X-rays to next-generation techniques that enable them to take high-resolution, information-rich images inside patients.
Still, surveying internal structures is just one element of the diagnostic process. Doctors still need to interpret what they see and cross-reference these details with databases depicting what’s considered normal to make a definitive diagnosis. Given the sheer number of features that need to be considered before making an accurate diagnosis, this process takes a long time and requires a wealth of experience. What if the process could be streamlined and accelerated with a helping hand from artificial intelligence?
"The analysis of three-dimensional imaging processes is very complicated," explained Oliver Schoppe, the first author of a study published by researchers at the Technical University of Munich, which aimed to address this challenge. Schoppe and colleagues have created a self-learning algorithm for analyzing and interpreting the vast amounts of data within medical images, which will ultimately give clinical diagnostic workflows a much-needed boost. The study was published in Nature Communications.
The platform, named AI-based Mouse Organ Segmentation (AIMOS), is powered by artificial neural networks (ANNs), computing systems inspired by the brain’s ability to learn new information. ANNs are organized into nodes, connected in a manner similar to how neurons “talk” to each other via synapses. These nodes are layered, with information traveling from the input to the output layers after traversing nodes within the entire network.
"You used to have to tell computer programs exactly what you wanted them to do," said Schoppe. "Neural networks don't need such instructions:" It's sufficient to train them by presenting a problem and a solution multiple times. Gradually, the algorithms start to recognize the relevant patterns and are able to find the right solutions themselves."
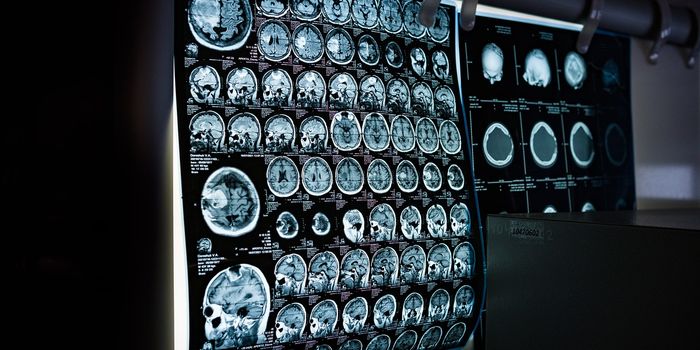
Here, the training regimen involved funneling images of 3D whole-body scans of mice into AIMOS. These images, obtained using high-powered fluorescence microscopy, showed internal organs, including the stomach, brain, spleen, liver, and kidneys, in stunning clarity and resolution: the distance between individual measurements was just six micrometers or the diameter of a single cell.
"We were lucky enough to have access to several hundred images of mice from a different research project, all of which had already been interpreted by two biologists," commented Schoppe.
AIMOS demonstrated its phenomenal capacity for learning in the speed with which it could interpret these high-definition images. "We only needed around ten whole-body scans before the software was able to successfully analyze the image data on its own - and within a matter of seconds,” said Schoppe.
“It takes a human hours to do this."
To validate the robustness and reliability of AIMOS, Schoppe and the team input an additional 200 mouse scans and observed that the system consistently outperformed human capabilities in terms of both speed and accuracy.
As this AI platform is being pushed towards medical diagnostic applications, it also has the capacity to drive efficiency in lab-based research protocols. "Images of mice are vital for, for example, investigating the effects of new medication before they are given to humans. Using self-learning algorithms to analyze image data in the future will save a lot of time in the future," said study author Bjoern Menze, head of the Image-Based Biomedical Modeling group.
Sources: Nature Communications, Technical University of Munich.