A group at the Icahn School of Medicine at Mount Sinai in New York City recently published a very interesting and exciting paper in
Nature Biotechnology detailing their computer model to predict how drugs affect the immune system.
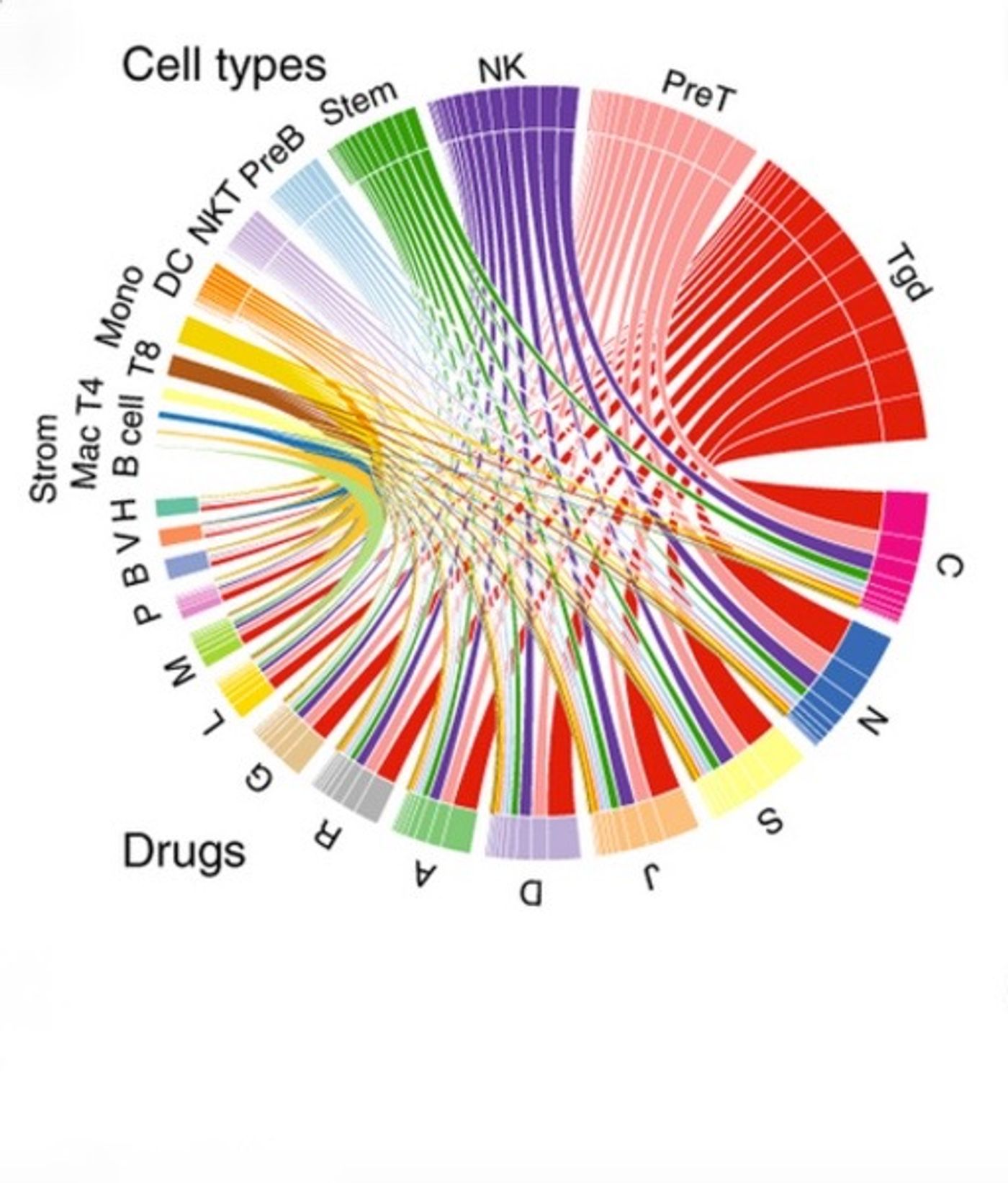
The strategy behind this model was to create a profile of targets that a drug engages with (chemogenomic profile) and a profile of the various gene expression states an immune cell could be in (immunogenomic profile). In total, the researchers categorized 304 state changes in 221 immune cell types that were then matrixed with 1,309 different drugs to create a list of 397,936 possible drug-immune cell interactions. Using the 69,995 significant interactions from this list, they created an immune cell pharmacology (IP) map. One large caveat to this paper is that the immune cell state change data was generated from mice and the drug data was gathered from humans. It was unclear if integration of these two data sets from two different species could provide any useful results. This is where the validation becomes incredibly important.
In order to prove that their IP map predictions are actually relevant to humans, the researchers compared their map with data from two independent patient populations. The IP map predicts that the general anesthetic propofol will increase neutrophils (an abundant type of white blood cell) and that spironolactone, an anti-hypertensive drug, will increase monocytes (another type of white blood cell). The electronic medical records of patients who had received either of these drugs and had blood cell counts collected within a month of receiving that drug were pulled out from the 2.3 million electronic medical records in the Mount Sinai Hospital System and evaluated. The shifts in the neutrophil or monocyte populations found in the patient data were significant and matched the shifts predicted by the IP map. To confirm this even further, the same procedure was done on electronic medical records from Columbia University Medical Center, with the same results.
In vivo experimental validation of the IP map also checks out. The researchers compared the effects of clioquinol and amantadine on neutrophil migration from the bone marrow to the blood. Clioquinol was predicted by the IP map to have a large effect on neutrophil migration and amantadine was predicted to have no effect, and that is exactly what the researchers found when they injected wild-type mice with the drugs.
The IP map has already proven useful. Experimental confirmation of the IP map prediction that guanfacine, a drug commonly prescribed for ADHD, increases the proportion of regulatory T cells means that this drug, which has already gone through safety and tolerability trials and has FDA approval, could also be used to promote peripheral tolerance. This is a perfect example of the value of this kind of technology. Repurposing drugs and getting them approved for other indications is an easy way for pharmaceutical companies to increase their revenue while bettering the care of everyone else by increasing their therapeutic options. It’s a real win-win. The IP map has other uses as well, with the authors citing its potential to be used as a tool to study immune cells in cancer and their response to various drug combinations. This paper is a fabulous example of how computer modeling can be used for clinical medicine and drug development. It is tools like this that are going to vastly improve patient care in the future.