Researchers have developed a novel way to determine stem cell fate earlier than current techniques allow. Stem cells have a special ability to turn into – differentiate – into any type of specialized cell in the body. Knowing what type of cell a stem cell will eventually differentiate into and the cell’s stage of differentiation will be a major benefit in the drive to use stem cell for therapeutic applications. The work has been reported in the open access journal
Scientific Reports.
The method is both straightforward and versatile, commented the senior author of the work, Prabhas V. Moghe, Distinguished Professor of Biomedical Engineering and Chemical and Biochemical Engineering at Rutgers. "It will usher in the next wave of studies and findings," he said.
Current techniques that check the growth stage of stem cells look at populations of cells as a whole and are not sensitive enough to determine the fate of specific cells. However, clinical use of stem cells, such as in bone marrow transplants after cancer treatment, require confidence that every implanted cell will grow into the desired cell type. Protein markers that are used to assay stem cell fate usually only show up on the cell after some differentiation, making them too late for clinical applications as well.
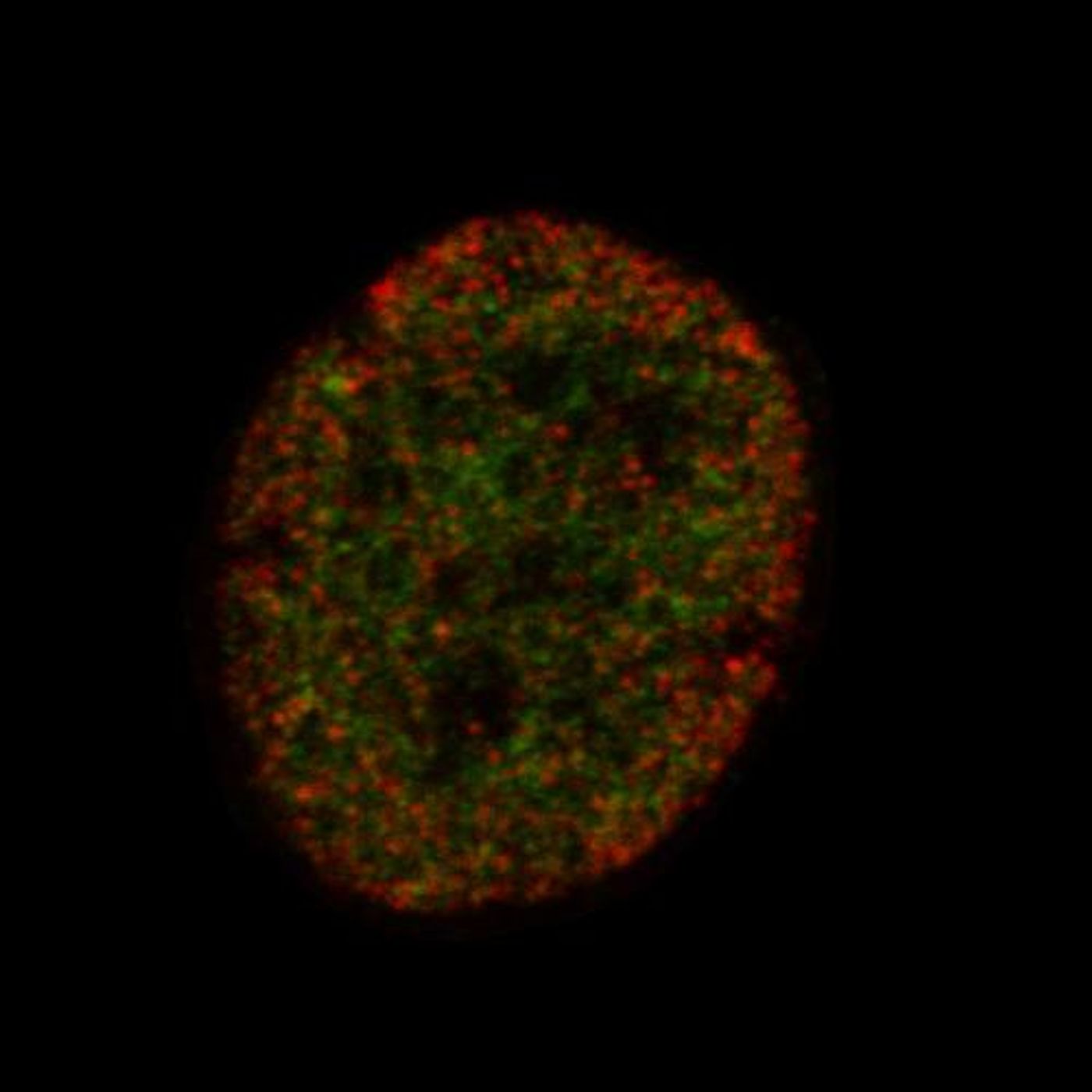
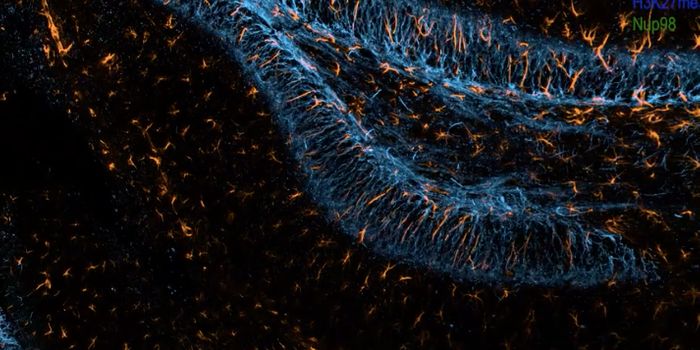
In order to find another way to learn the fate of a stem cell, a research collaboration between a variety of fields in multiple universities utilized super-resolution microscopy to look at epigenetic modifications. Those modifications can alter the structural conformation of DNA and change the genes that are being turned on and off. Some of those modifications indicate the transition of a stem cell into a specific cell type like a bone or fat cell. This tool enabled researchers to learn the fate of a stem cell far earlier than with other techniques.
This method has been termed EDICTS for Epi-mark Descriptor Imaging of Cell Transitional States; the epigenetic modifications made to genes are labeled, then super-resolution imaging is used to visualize their exact locations.
"We're able to demarcate and catch changes in these cells that are actually not distinguished by established techniques such as mass spectrometry," Moghe said. He described the method as "fingerprinting the guts of the cell," and the results are quantifiable descriptors of each cell's organization (for example, how particular modifications are distributed throughout the nuclei).
Their method was experimentally confirmed by assaying two epigenetic modifications in human stem cells that were grown in the lab. After exposing the cells to chemicals stimulating different kinds of differentiation – to fat and bone cells, with another as a control – the modifications had changed location in cells transitioning to different cell types. Those changes were enough to tell the difference between the individual cells as well.
"The levels are not significantly different, but how they're organized is different and that seems to correlate with the fact that these cells are actually exhibiting different fates," said Moghe. "It allows us to take out a single cell from a population of dissimilar cells," which can help researchers select particular cells for different stem cell applications. He added that this tool is as simple as labeling, staining and visualizing; a familiar procedure for many investigators. Here, the critical piece of the puzzle is an imager capable of imaging at super-resolution, which allows the procedure to be widely used.
If you would like to know more about stem cell differentiation and fate, check out the following video featuring David Schaffer, a Professor of Chemical and Biomolecular Engineering, Bioengineering, and Neuroscience at the University of California, Berkeley, and the Director of the Berkeley Stem Cell Center.