Artificial Intelligence for Cell Analysis in Biologics Development
There’s No Turning Back
Not long ago, solving the crystal structure of a protein required an entire PhD.
Growing crystals, collecting X-ray diffraction data, and interpreting electron density maps often took years of optimization and expensive instruments. Even then, solving all protein structures was a challenge, further compounding the “protein folding problem” in biology.
Fast forward to today – Google DeepMind’s AlphaFold 3, powered by AI, can predict the three-dimensional structures of proteins, DNA, RNA, and ligands in minutes with incredible accuracy. Scientists worldwide can access these predictions on the free AlphaFold Server, accelerating scientific progress.
AI’s impact is visible across all areas of drug research and development, from protein optimization to cell analysis, helping labs maximize efficiencies.
Biologics R&D in the Age of AI
AI-driven computational methods are revolutionizing the de novo design of single-domain antibodies and miniproteins, significantly speeding up the of new biologics and even targeting the “difficult to drug” space, like G protein-coupled receptors (GPCRs) and ion channels.
The integration of AI in biologics development is supported by the growing contributions of digital solutions providers. Companies like Svitla Systems offer outsource programming services and custom software development, enabling advancements in cloud technologies and AI applications. These technological capabilities play a role in streamlining complex processes across sectors, including healthcare and drug discovery.
For instance, researchers can use AlphaFold 3 to predict the structure of an antibody and its binding affinity to a target antigen. This modeling optimizes antibody design while reducing the time and resources needed for experimental testing.
NVIDIA, a leading technology company, has expanded its offerings to include AI and deep learning solutions with vast applications in biosciences.
Amgen, a prominent biotechnology company, is cutting the time and cost of biologics development by using NVIDIA's AI and computing capabilities to predict protein structures, optimize molecular interactions, and speed up drug discovery processes.
The “Cell Analysis Problem”
Cell imaging and analysis are critical in research workflows for assessing cell health, viability, and proliferation, ensuring that only healthy cells are used in experiments. In biologics, high-throughput cell assays are routinely used to determine the fitness of antibody-producer cell lines or measure cellular responses to biotherapeutic candidates.
Before advanced tools were available, analyzing cell images was a manual job requiring hours of work by experts with a keen eye. Besides the obvious throughput disadvantage, such analysis is subjective and prone to error.
Another concern is the biological relevance of cell data. Like most living things, cells show their true nature when unperturbed. But the labeling and imaging protocols used to visually track cell behavior can influence cell biology and the resulting data, especially with sensitive cell types like stem cells.
Jasmine Trigg, a Scientist at Sartorius, agrees. “Before modern tools, I had a hard time analyzing what I needed. It was tough to accurately segment cells or analyze them on a larger scale. Another challenge was using imaging reagents, especially with neural cells. Sometimes, they weren’t compatible with sensitive cells or introduced artifacts,” Trigg recalls from her days in a neuroscience lab.
Scientists like Trigg are addressing these issues with the next generation of live-cell analysis tools paired with AI-enabled software.
AI-Enabled Live-Cell Analysis
Modern instruments for live-cell analysis provide high-quality insights much faster. First, these technologies are non-disruptive by design. For example, the Incucyte® Live-Cell Analysis System houses the microscope and imaging functions within a cell culture incubator, so you can monitor the cells in real time without moving them.
Second, AI-enabled software enhances these tools by automating the tedious process of analyzing thousands of cell images in high-throughput applications.
These software modules rely on deep learning methods trained on extensive datasets of cell images. High-quality training datasets are crucial because they provide the foundation for the AI to learn various cell types, conditions, and anomalies.
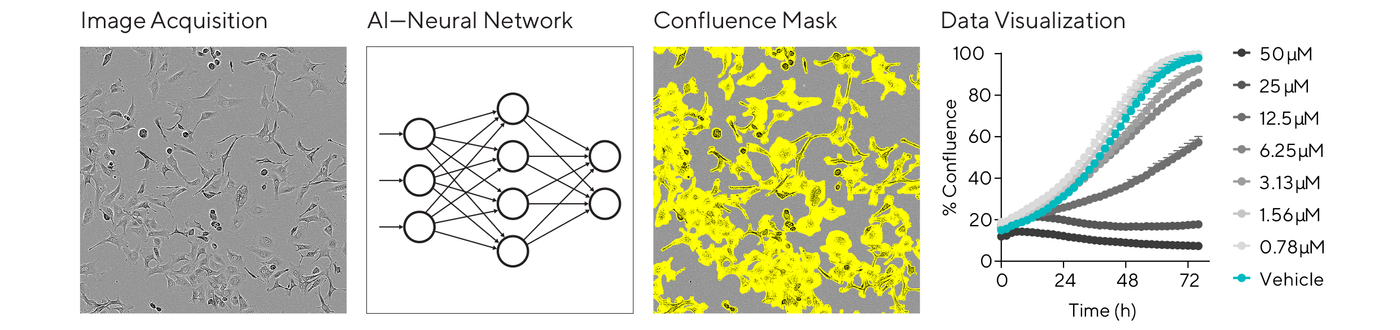
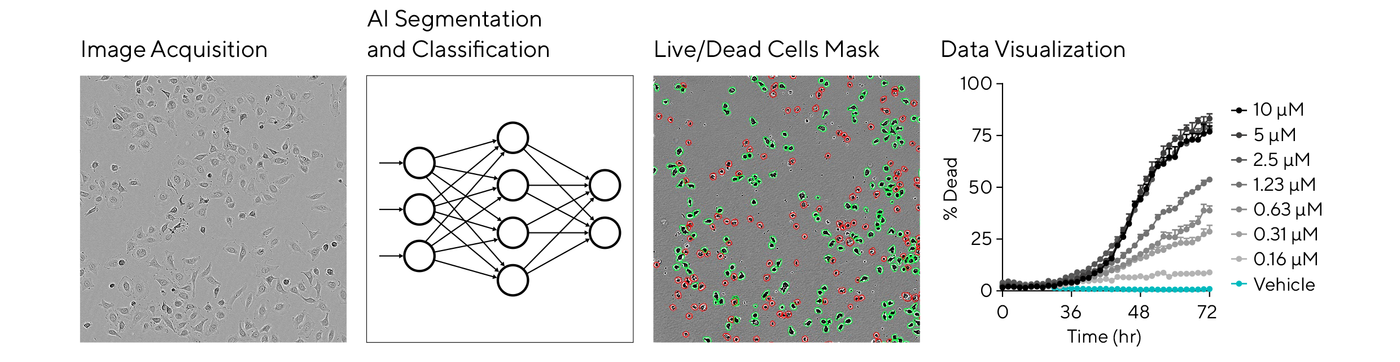
A typical analysis workflow starts with segmentation, delineating individual cells from the background, followed by classification to identify cell types, states, or specific features of interest. This automation and objective analysis are key to providing consistent and reproducible results in dose-response assays or cell optimization studies.
Label-Free Cell Confluence and Viability
An important decision when designing cell assays is whether to track events with or without fluorescent labeling. Fluorescence has many practical benefits in cell analysis, but sometimes it’s not an option. Researchers working with complex disease models or patient-derived cells are increasingly choosing non-disruptive, label-free methods.
Advancements in AI are providing powerful analysis capabilities for basic measurements of proliferation and cell death without using labels. For instance, the basic analysis software of the Incucyte® system includes an AI Confluence algorithm for automatically segmenting phase contrast cell images with minimal user input. Backed by a neural network trained on many classes of cells, this workflow is simpler, more objective, and repeatable, scalable to a lab’s throughput needs.
For label-free cell health analysis on the Incucyte® system, the AI Cell Health Analysis module uses AI-driven algorithms to kinetically quantify live and dead cells based on trained neural networks. Watch this new webinar to hear more from Jasmine Trigg on the challenges in cell health analysis and the latest AI tools for live-cell imaging.