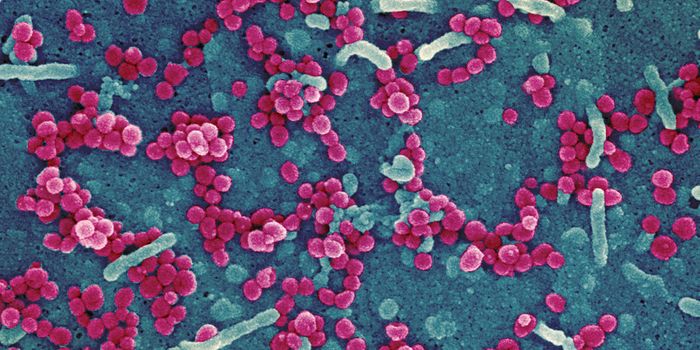
The Genetic Hotspots That Can Lead to Cancer
The genetic material carried by cells in our body has to replicate every time a cell divides, so for some cell types, the DNA undergoes many rounds of replication. There are some genetic hotspots, in which stability is low, and the risk of mutations is high. It has been shown that genetic mutations that occur during regular cell division are associated with the development of cancer. Scientists at Indiana University have aimed to learn more about these genetic hotspots, hopefully, to inform our understanding of cancer development. This work used a bacterium, Escherichia coli, but the basics of the work is applicable to human systems.
"This research gets us closer to understanding how the cell's replication machinery interacts with DNA," said Patricia Foster, a professor emerita in the IU Bloomington College of Arts and Sciences' Department of Biology and one author of this work, in Genetics. "If you can understand exactly why an error occurs at a particular point on the DNA in bacteria, it gets you closer to understanding the general principles." (A second Genetics paper on this data was also published.)
Some tissues reproduce new cells frequently, so there is a higher risk of mutations happening in that DNA. The cellular proofreading machinery can’t catch every mutation, so a person may end up with a genetic mutation even though they are not born with one, called a somatic mutation. The E. coli bacterium also undergoes many rounds of replication, making it a good model for this work.
"There are parts of the genome that contain 'cancer drivers,' where changes in the DNA can allow tumor cells to proliferate," explained Foster. "If you could know what sections of the DNA had a higher risk for mutation, you might be able to focus your analysis on these 'hotspots' to predict what will happen next."
The chance of errors, or mutations, in genetic replication, were around eighteen times higher in places where one of the four nucleotide bases that make up DNA repeated multiple times in a row. Those mutations were as many as twelve times as likely in sequences with a three-base pattern.
Although it’s been known that these patterns are more likely to experience errors, this data was created with statistical power. These error rates are purported to be far more accurate than other estimates.
DNA proofreading does still do an essential job. There is an enzyme that can perform that function, and there is a cellular pathway, called mismatch repair. When the scientists turned the proofreading enzyme off, there were 4,000 times as many mutations. Mismatch repair serves as a backup for that enzyme, and when that was also switched off, there were another 200 times as many errors.
"When we switch off these backup systems, we start to see 'pure' errors -- the places where the polymerase is more likely to make a mistake without intervention from other processes, " Foster said. "Until now, I don't think anyone could truly see the seriousness of these error hotspots in DNA."
To learn more about how somatic mutations and gremlin mutations differ, and how that impacts cancer, check out the video from the Dana Farber Cancer Institute.
Sources: AAAS/Eurekalert! Via News at IU Bloomington, Genetics 2018 Niccum et al, Genetics 2018 Foster et al