Scientists Engineer a Better Way to Direct Evolution
Earlier this year, three researchers shared the Nobel Prize in Chemistry for pioneering a process called directed evolution, in which diversity is introduced into a genome with the aim of creating a new biomolecule. Those new biomolecules have a wide array of potential applications, especially in drug development. The process was considered painstaking and laborious; however, scientists at the University of California, Irvine have now streamlined the process. In work reported in Cell, they have used live yeast cells as a conduit to accelerate genetic mutations, which can be introduced stably as the yeast reproduces and evolves.
“By moving high rates of diversification into cells in a targeted manner, we can grow and pressure those cells to evolve into something new from any genes of our choosing,” explained first author Arjun Ravikumar, a recent recipient of a biomedical engineering Ph.D. at UCI. "Our work has reduced evolution to be an extremely rapid, straightforward and scalable process."
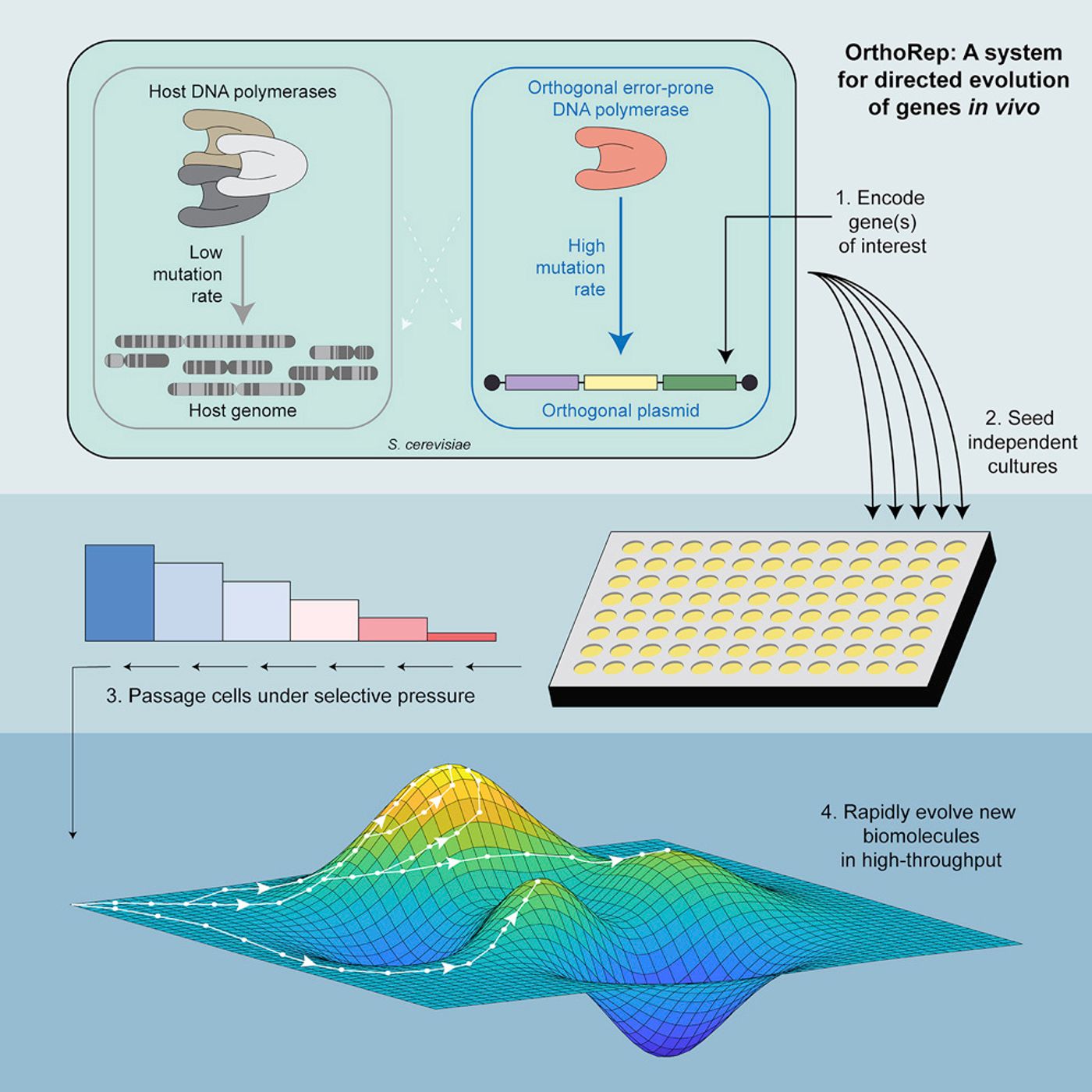
Introducing changes into a genome can be a challenging prospect. In this work, the researchers created a molecular system that could introduce errors in live yeast cells. Called OrthoRep, the "DNA polymerase-plasmid pair" creates mutations in a host cell's genome about 100,000 times faster than the natural rate. Their method also allows for researchers to see what is happening in the genome as evolution happens.
The number of evolutionary cycles the genome goes through is very important when using directed evolution, noted the senior author of this study, Chang Liu, UCI assistant professor of biomedical engineering. Every one of those cycles may add a new function or improve an existing one. “But if each cycle requires repetitious test-tube DNA molecular biology processing, you can only reasonably go through a few iterations,” he said.
"In contrast, natural evolution runs cycles continuously, essentially by culturing cells over time in an environment that pressures them to develop some new function; the problem from a biomolecular engineering standpoint is that the process is very slow," Liu added. "We have figured out a genetic architecture that allows biomolecular evolution to be very fast."
Liu suggested that this new work will help investigators with experiments that were previously very challenging to perform. In their report, the UCI team did 90 replicates of an enzyme evolution experiment to look for all of the changes it made, in this case, to develop drug resistance.
"There are many ways of solving a particular evolutionary challenge such as drug resistance, so the ability to run evolution experiments at the scale that we have allows us to capture and understand more of those possibilities, giving us therapeutically relevant insights into how resistance arises," Liu said.
The team plans to continue their studies; Liu wants to use the new technique to create antibodies that can constantly evolve to fight disease, or to make enzymes that engineer better drugs.
"Instead of having to inject an antigen into an animal in order to isolate an antibody, imagine just putting it into a culture of yeast cells and having it come out as a specific antibody," he said. "That could revolutionize how these and other protein drugs are discovered and developed."
One of this year’s Nobel Prize winners, Frances Arnold commented, "Directed evolution is a powerful way to build new proteins, but it can certainly benefit from technological innovations. The technique that Professor Liu and Dr. Ravikumar have developed will stimulate new applications and new avenues of investigation which will continue to expand our ability to compose new DNA."
Arnold is featured in the video above, discussing directed evolution.
Sources: AAAS/Eurekalert! via University of California, Irvine, I & EC Research, Cell