How Non-Coding Genomic Regions Influence Autoimmune Disease
Genes code for proteins, which are the cornerstone of many biological processes. The genome holds the instructions for making them in protein-coding genes and controls their activity with regulatory regions. There's also a massive amount of sequence that does not code for protein and has no readily apparent function. Researchers have been seeking to learn more about these 'dark' regions of the genome that had once been dismissed as 'junk.' New research has suggested that susceptibility to complex autoimmune and allergic disorders may lie in some of these regions. The findings, which have been reported in Nature, could help open new therapeutic avenues for these diseases.
Genome-wide association studies have enabled researchers to learn more about how small differences in the genome can contribute to a person's risk of various complex diseases (explained in the video at the end of the article). Susceptibility to many autoimmune diseases has been traced to a region of chromosome 11, some of which do not code for proteins. We don't really know how to manipulate these non-coding genomic regions with drugs that often target proteins in some way. We do know that many of these risk factors are in genetic regions that are characterized as enhancers. While they don't code for proteins, enhancers can help regulate gene expression - the production of proteins.
Scientists found an enhancer that the immune system needs in order for regulatory T cells to mediate the immune response and maintain balance.
"The immune system needs a way of preventing reactions to harmless self- and foreign substances and Treg cells play a vital role in this. They're also crucial in maintaining balance in the immune system so that our immune responses are kept in check during infections. Tregs only represent a small percentage of the cells making up our complete immune system but they're essential; without them we die from excessive inflammation," explained lead researcher and Babraham Institute group leader, Dr. Rahul Roychoudhuri.
"Despite this important role, there has been little evidence that unequivocally links the genetic variations that cause certain individuals to be susceptible to inflammatory diseases to changes in Treg function. It turns out that non-protein-coding regions provided us with the opportunity to address this important question in the field," noted Roychoudhuri.
By studying genomic regions that were not only conserved between species, but also among sections of the genome, the researchers found an enhancer region in mice that is analogous to the human region.
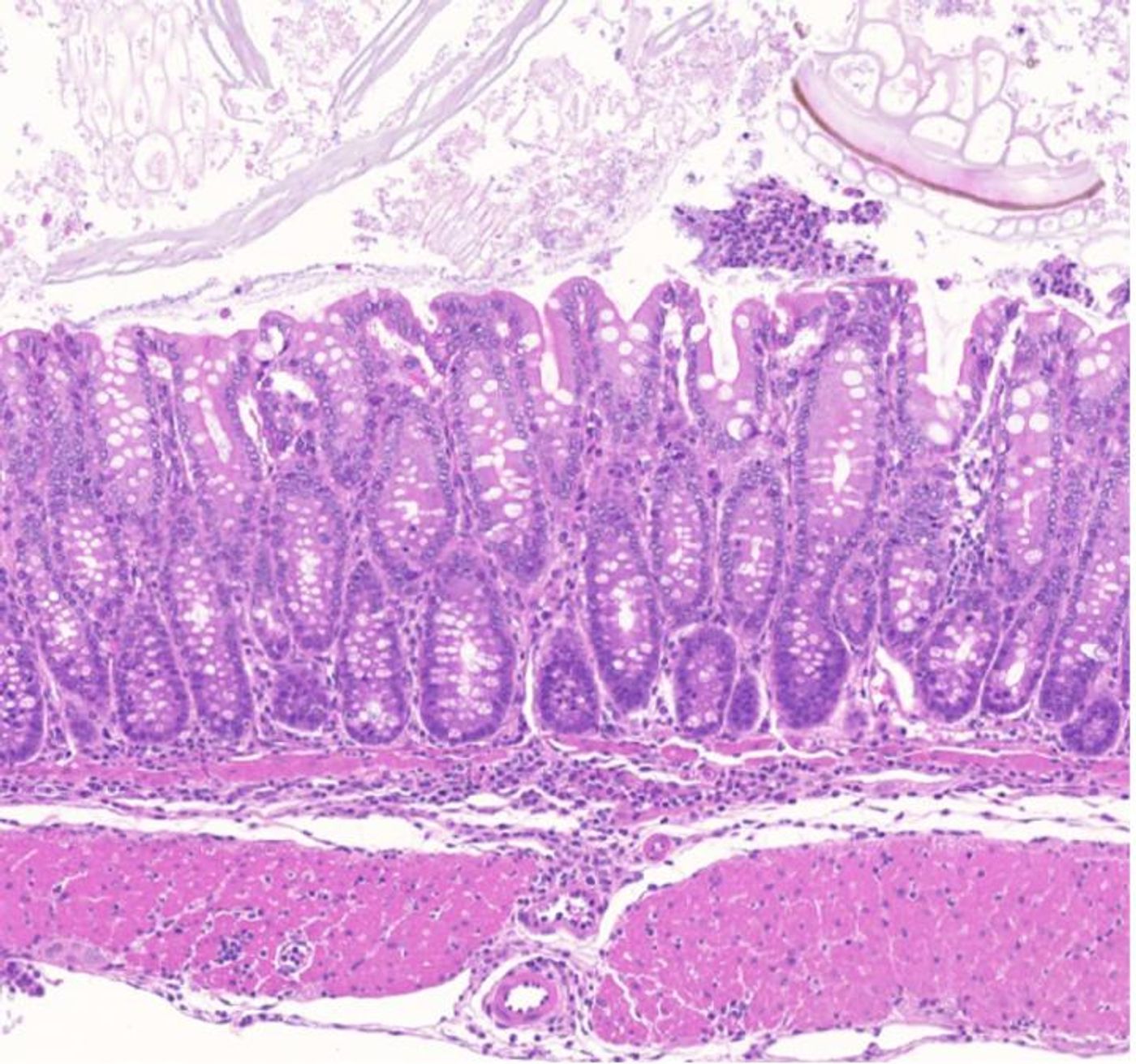
In the mouse, the scientists found that an enhancer helps regulate a Treg gene called GARP (Glycoprotein A Repetitions Predominant). When the enhancer was deleted, the GARP protein was lost from Tregs, and colon inflammation was out of control.
"Genetic variation provides important clues into disease processes that can be targeted by drugs. In our joint efforts here, we combined human and mouse research to gain invaluable insight into complex processes underlying immune diseases. This has identified GARP as a promising new drug target and brings us a step closer to developing more efficient therapies for people suffering from diseases such as asthma or inflammatory bowel disease," said the senior study author Dr. Gosia Trynka of the Wellcome Sanger Institute and Open Targets.
"Decades of research have now identified the variations in our genomes that make some of us more susceptible to inflammatory diseases than others. It has been very difficult, however, to make sense of how these variations relate to immune disease since many of them occur in non-protein-coding regions, and therefore the implications of these changes are poorly understood," said Roychoudhuri. "Studies such as these will enable us to link the genetic switches that commonly reside in such disease-associated non-coding regions with the genes they control in different cell types. This will yield new insights into the cell types and genes underlying disease biology and provide new targets for therapeutic development."
Sources: AAAS/Eurekalert! via Babraham Institute, Nature