How CRISPR Can Aid in Wildlife Conservation
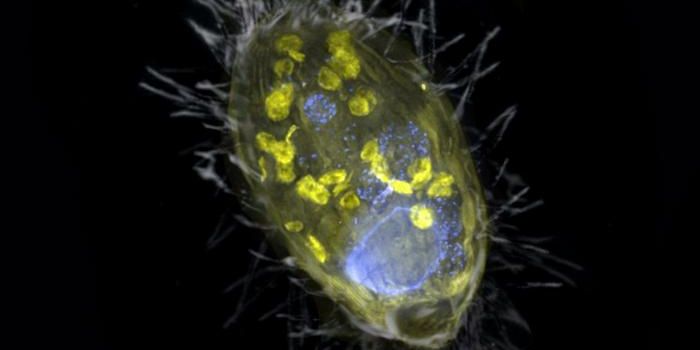
Since it was developed, researchers have modified and applied the CRISPR gene-editing technology in many different ways. In one of the latest applications of the tool, researchers have found a way to use it to differentiate between species in the field. Reporting in Molecular Ecology Resources, researchers used a CRISPR detection system called SHERLOCK to identify threatened species of fish (in this case, endangered Delta smelt) in real-time, without extracting any DNA. The researchers are hopeful that this technique will have a major impact on environmental monitoring.
"CRISPR can do a lot more than edit genomes," said study co-author Andrea Schreier, an adjunct assistant professor in the UC Davis animal science department. "It can be used for some really cool ecological applications, and we're just now exploring that."
This research concentrated on three species of fish that call the San Francisco Estuary home: the Delta smelt (threatened in the US and endangered in California), the longfin smelt (threatened in California), and the nonnative species wakasagi (sometimes referred to as freshwater smelt). It is notoriously challenging to specify which of these species a particular fish is, especially at younger stages.
"When you're trying to identify an endangered species, getting it wrong is a big deal," said lead author Melinda Baerwald, who was a project scientist at UC Davis when the study was developed, and is now an environmental program manager with the California Department of Water Resources.
Some pumping projects have to be reduced if too many endangered fish get sucked into the pumps, so it's important to rapidly differentiate between the species. Usually, scientists swab the fish or clip the fin, then extract the DNA in a lab and perform genetic sequencing, which all takes about four hours, without travel time, in the best-case scenario.
SHERLOCK (Specific High-sensitivity Enzymatic Reporter Unlocking) shortens the process dramatically, to a test that only takes about 20 minutes and can be done virtually anywhere. A flow strip like what's used in a urine test or a fluorescent detector can be used to identify the species quickly.
"Anyone working anywhere could use this tool to quickly come up with a species identification," Schreier said.
The video above describes how SHERLOCK is used in diagnostics.
More research will be needed to determine whether this method will work for other organisms, but the researchers expect that it can be used for other kinds of identification assays.
"There are a lot of cryptic species we can't accurately identify with our naked eye," Baerwald said. "Our partners at MIT are really interested in pathogen detection for humans. We're interested in pathogen detection for animals as well as using the tool for other conservation issues."
Sources: AAAS/Eurekalert! via University of California - Davis, Molecular Ecology Resources