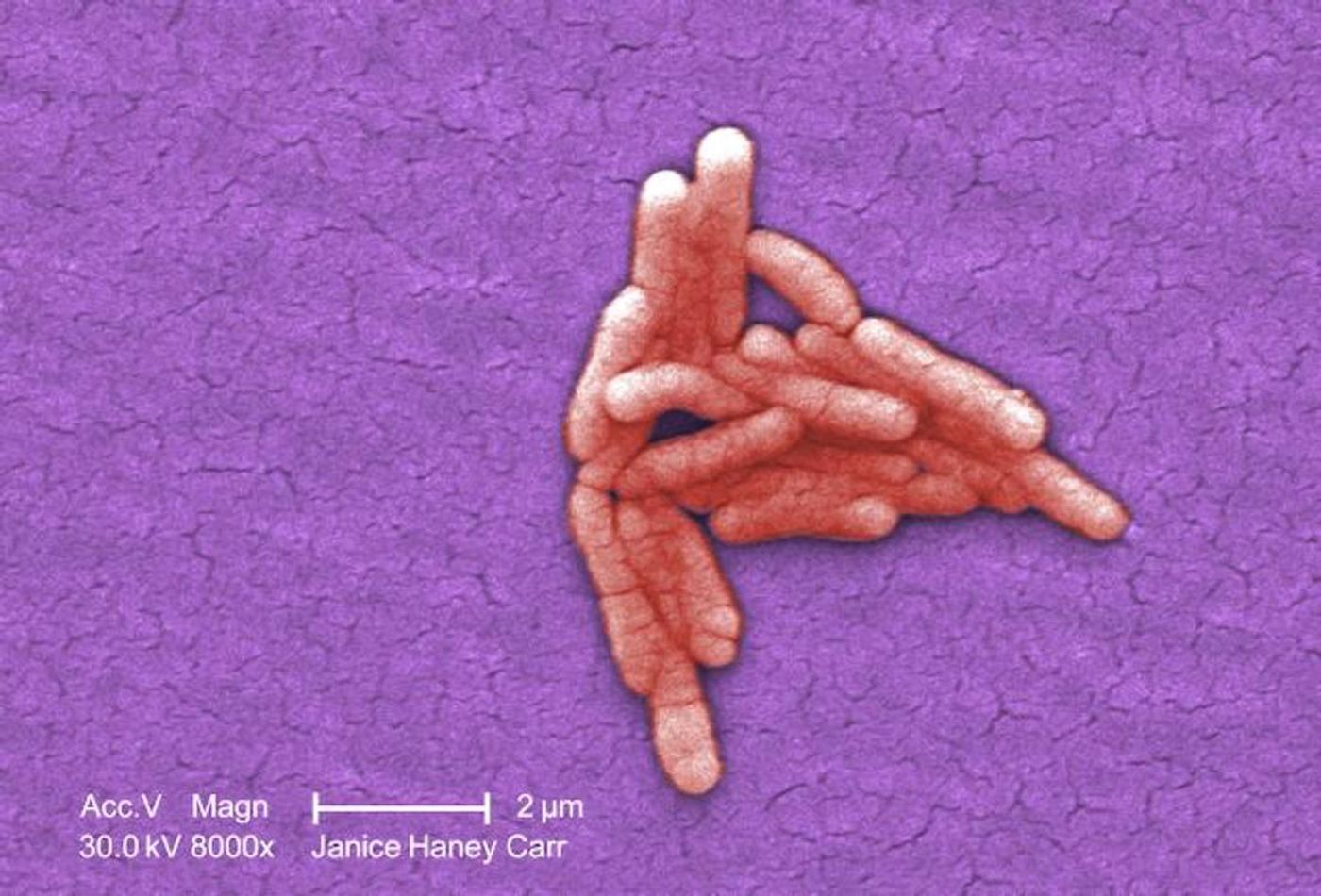
Tracing the Evolution of a Deadly Microbe
Salmonella bacteria are human and animal pathogens that are found everywhere. When this bacterium infects a person, it causes an illness called salmonellosis, which affects millions of people around the world every year. These illnesses typically cause gastrointestinal problems but if the infection progresses to the bloodstream, it can become a serious disease that needs to be treated with antibiotics, and may result in death.
Scientists have now learned more about how one strain of Salmonella bacteria found in Africa evolved to become deadly. The strain, called Salmonella Typhimurium ST313, has killed hundreds of thousands of people in Africa who are immunocompromised. It’s thought to be responsible for the deaths of about 50,000 people in Africa every year.
Reporting in Nature Microbiology, an international team of researchers analyzed samples that were collected from patients infected with Salmonella from 1966 to 2018. They recovered 680 isolates, and analyzed the genetic sequences of the pathogens. This enabled them to generate a timeline of Salmonella evolution that showed how the microbe changed over many years.
The scientists were able to identify genetic mutations in the bacteria that made some strains more invasive than others, as well as ‘degradation events’ in which the microbe acquired mutations that caused the loss of some of its functions.
This work has also led to the discovery of a new strain of ST313 that is susceptible to antibiotics. The study authors suggested that this susceptible strain may have emerged when clinics in Malawi stopped using a particular antibiotic called chloramphenicol to treat infections.
"Through a remarkable team effort we have removed some of the mystery about the evolution of African Salmonella. We hope that by learning how these pathogens became able to infect the human bloodstream we will be better prepared to tackle future bacterial epidemics,” said the study leader Professor Jay Hinton at the University of Liverpool.
Hinton has been studying Salmonella for many years, and is the leader of an effort to understand the virulence, transmission, and epidemiology of invasive non-Typhoidal Salmonellosis, called the 10,000 Salmonella Genomes Project.
"By combining the power of genomic analysis with epidemiology, clinical observations, and functional insights, we have shown the value of using an integrated approach to link scientific research with public health,” said study author Dr. Caisey Pulford.
Sources: AAAS/Eurekalert! via University of Liverpool, Nature Microbiology




![Everything You Need To Know About NGS [eBook]](https://d3bkbkx82g74b8.cloudfront.net/eyJidWNrZXQiOiJsYWJyb290cy1pbWFnZXMiLCJrZXkiOiJjb250ZW50X2FydGljbGVfcHJvZmlsZV9pbWFnZV9mNTM1ZjIyYzA5MDE5ZmNmMWU5NmI0ZDc4NWU2MzdiZTZlN2I5ZDk5XzE4NDUuanBnIiwiZWRpdHMiOnsidG9Gb3JtYXQiOiJqcGciLCJyZXNpemUiOnsid2lkdGgiOjcwMCwiaGVpZ2h0IjozNTAsImZpdCI6ImNvdmVyIiwicG9zaXRpb24iOiJjZW50ZXIiLCJiYWNrZ3JvdW5kIjoiI2ZmZiJ9LCJmbGF0dGVuIjp7ImJhY2tncm91bmQiOiIjZmZmIn19fQ==)