Sequencing and Identifying SARS-CoV-2 Variants Efficiently
Viral surveillance can give researchers vital information about virus origins, transmission routes, affected populations, and mutation and evolution rates. This can be used to design and evaluate diagnostic tests to track virus infection rates. This type of genomic surveillance information can be generated using next-generation sequencing (NGS). NGS methods include whole-genome sequencing (WGS) as well as more targeted approaches to enrich regions of interest via hybridization capture or multiplex PCR. NGS methods can be used for wastewater-based epidemiology, genomic epidemiology, and variant calling in addition to viral surveillance.
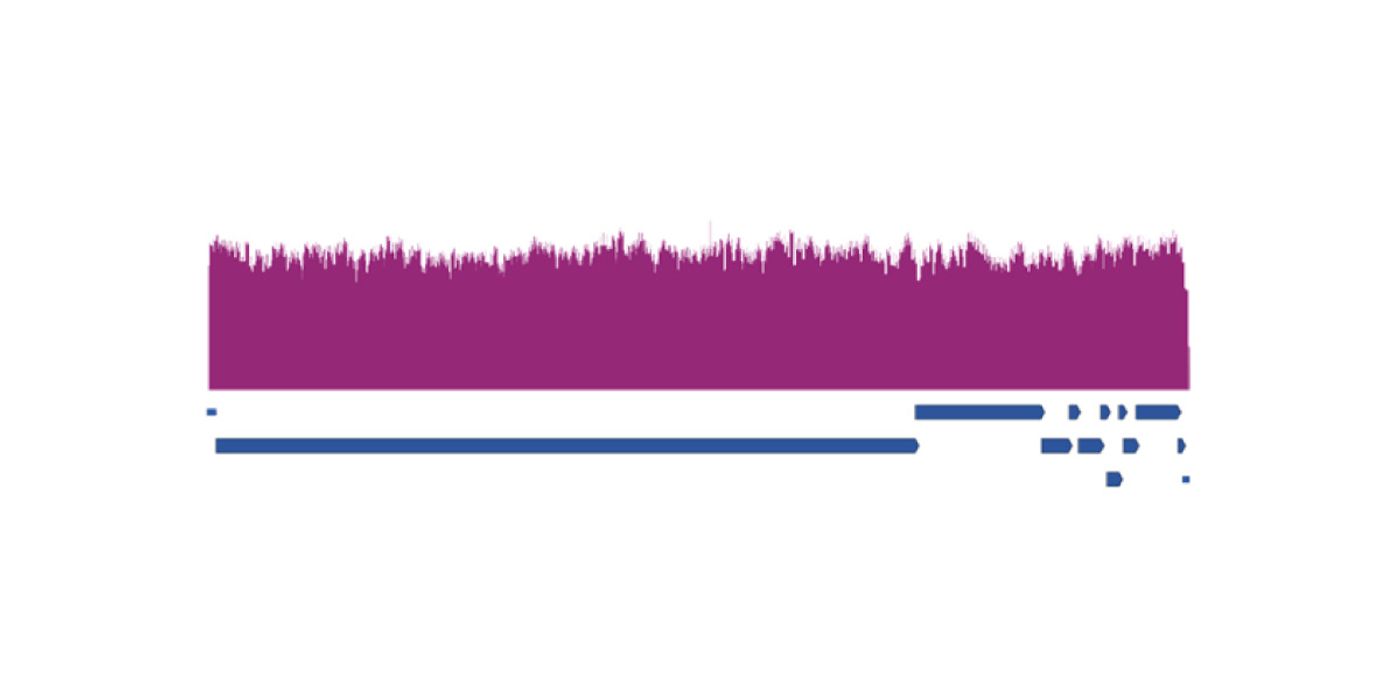
New SARS-CoV-2 variants are being discovered at alarming rates. In response, NGS workflows have been established to identify and characterize new strains. The ARTIC network, a collaborative project to progress molecular epidemiology, has made available an NGS panel based on amplicon sequencing; it’s called the ARTIC v3 Multiplex PCR Primer Assay. Amplicon sequencing is a type of targeted next-generation sequencing that uses PCR to create sequences of DNA called amplicons. “The ARTIC nCov-2019 amplicon sequencing protocol has been widely adopted across the world, and the genome data is critical to understanding and tracking the outbreak,” says Dr. Josh Quick, a researcher at the University of Birmingham who was an early user of the ARTIC network.
Another amplicon enrichment assay, the Midnight Multiplex PCR Primer Assay, was developed by Drs. Nikki Freed and Olin Silander of Massey University in New Zealand. The Midnight Panel generates fewer, but much longer, amplicons than the ARTIC Panel, which can allow for more even sequencing coverage. When paired with the IDT Lotus™ DNA Library Prep Kit, internal benchmarking studies of both the ARTIC and Midnight Multiplex PCR Primer Assays show complete SARS-CoV-2 genome coverage. Additionally, they can both be used to sequence the S-gene, a commonly mutated gene that contributes to SARS-CoV-2 infectivity and can be used as an indicator of viral evolution. For more information on the ARTIC and Midnight protocols, visit our website.
One weakness of these multiplex PCR assays is that mutations in the sequences of the virus that correspond to PCR primers can cause dropout in virus sequencing. All current SARS-CoV-2 strains can be identified using the ARTIC primer sets, though some sets cause gaps in sequence coverage. Longer amplicon sizes generated by the Midnight assay mean that fewer primer sets are needed to amplify the viral genome; this makes Midnight less likely to be disrupted by mutations.
The Swift Normalase™ Amplicon Panel (SNAP) also applies amplicon sequencing to viral surveillance. The SNAP SARS-CoV-2 Multiplex Assay leverages Swift’s patented multiplex PCR technology, enabling library construction from cDNA using overlapping primer pairs to target 99.7% of the SARS-CoV-2 genome with a single pool of multiplexed primer pairs. The SNAP assay is optimized for short-read sequencing but is also compatible with low-input and damaged samples, including wastewater, and complete S-gene coverage is also available. The SNAP protocol takes half the time of the ARTIC and Midnight protocols (3 hours vs. 6+ hours) and is performed in a single tube, creating overlapping amplicons. This technology results in built-in redundancy, which can provide coverage in the case of possible primer dropout to maintain a high level of genomic coverage, allowing variant discovery. Adriana Heguy, PhD, from Langone Health at the NYU Grossman School of Medicine, said that “Swift has been a valued research partner, and we look forward to working with them to continually improve the ability of amplicon-based methods to achieve greater coverage in fewer reads, which would enable us to achieve good genome coverage for low-viral-load samples.” For more information about Swift’s multiplex PCR technology or the SNAP SARS-CoV-2 Assay, please check out our webinar. Title: Efficient and scalable amplicon sequencing solutions for SARS-CoV-2 surveillance
RUO - For research use only. Not for use in diagnostic procedures. Unless otherwise agreed to in writing, IDT does not intend these products to be used in clinical applications and does not warrant their fitness or suitability for any clinical diagnostic use. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations.