CGP: Simplifying the path from tumor to treatment
The promise of NGS in personalized cancer care
For years, next-generation sequencing (NGS) has enabled oncologists to identify DNA mutations that are associated with the onset, growth, and metastasis of individual tumors—empowering them to select treatments that are most likely to be effective and extending patients’ lives. This type of information-driven treatment is one of the hallmarks of “personalized medicine.”
NGS of tumor samples can also provide guidance on whether common treatments might be ineffective in particular cases, giving oncologists the ability to rapidly change treatment plans and reduce patient suffering by avoiding unnecessary side effects.
One such example is a type of driver mutation common in non-small cell lung cancer (NSCLC). Many of these tumors contain activating mutations in the epidermal growth factor receptor (EGFR) gene, which can usually be targeted by EGFR tyrosine kinase inhibitors (TKIs); this blocks the activation of pathways leading to continued proliferation and metastasis. However, as many as 10% of EGFR mutations in NSCLC contain insertions in exon 20 (Ex20ins), which renders them resistant to these TKIs; identification of these variants can thus lead to more appropriate treatment.
This kind of data-driven decision making is a sharp contrast to pre-NGS screening methods, in which tumors were defined by their tissue of origin or by using serial tests that rely on PCR or immunohistochemistry. While these methods can show the absence, presence, or over-expression of specific genes or variants, and are still valuable in many contexts, they can’t compete with the nucleotide-level information provided by NGS.
NGS assays must evolve to keep pace with the expanding landscape of cancer genomics and treatment
Most NGS-based methods used in tumor profiling include the enrichment of known cancer-associated genomic regions. This “targeted NGS,” or “target-enriched NGS,” is much more efficient compared to whole-genome sequencing (WGS) due to the depth of sequencing required to identify rare alleles; targeted approaches significantly decrease the amount of sequencing required and the resources needed for data storage and analysis.
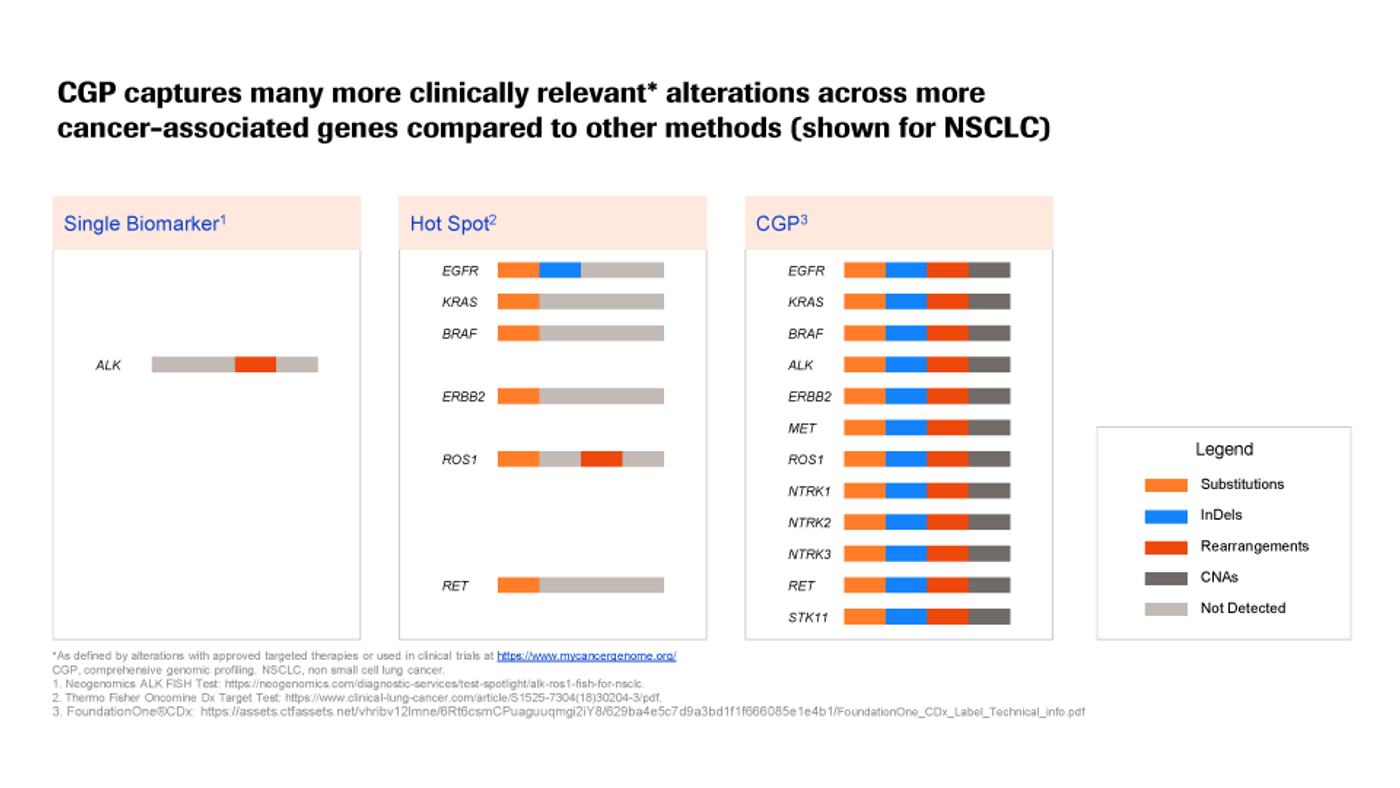
However, while targeted NGS is an incredibly powerful tool for cancer research and treatment, many methods focus on only a very narrow region of the genome—sometimes only one gene or a handful of genes (see Figure 1). Others, such as cancer hotspot assays and hereditary oncology assays, may cover more (up to 50 or ~200 genes, respectively)… but the total number of known cancer-associated variants spans a much greater and ever-growing portion of the genome; to date, there are several hundred clinically-actionable variants across hundreds of genes in dozens of tumor types.
Figure 1
This means that these highly-targeted methods may miss valuable, potentially life-saving information, or delay its discovery … and, with cancer, speed matters.
CGP strikes a balance, providing focused genomic insights into many more tumor types
This is where comprehensive genomic profiling, or CGP, comes in. There are several types of CGP assays, and while their technologies and specific targets vary, they share several advantages over other, smaller-scale targeted NGS methods:
- Simultaneous interrogation of hundreds of cancer-associated regions spanning dozens of cancer types
- Much-reduced likelihood of missing informative variants—both actionable variants and others, such as EGFR Ex20ins mutations, which may be critical for treatment decisions
- Detection of all major mutation classes, including single-nucleotide variants (SNVs), gene fusions and rearrangements, and copy number alterations (CNAs)
- Analysis of broader genomic signatures that are important indicators of tumor progression or treatment response (tumor mutational burden (TMB), microsatellite instability (MSI), and genomic loss of heterozygosity (gLOH))
- Creation of a comprehensive data set—from a single sample—which can be re-interrogated as new cancer biomarkers emerge and new treatments become available… potentially increasing opportunities for enrollment in clinical trials
In short, CGP panels enable comprehensive, detailed molecular tumor profiling from one sample and one test, provided in a single report—and offer the potential to accelerate and refine the path from biopsy to the most appropriate treatment, reducing suffering and extending lives.