New Digital Imaging Analysis Spots HIV Degradation
A new system that tracks proteins as they come and go in the cell is predicted to help scientists identify therapeutic targets for a multitude of diseases including HIV, cancer, and Alzheimer’s. From the Sanford-Burnham Prebys (SBP) Medical Discovery Institute, scientists focus first on HIV, using the new imaging-based approach to study protein stability.
The new “Global Arrayed Protein Stability Analysis" (GAPSA) system is the first cell-based platform that screens for the production and destruction of proteins via high-throughput, genome-scale imaging. It works by identifying circuits in proteins that drive destruction.
In the present study, researchers focused on identifying human proteins degraded by HIV as it promotes its infection process. But in addition to identifying HIV-associated proteins, GAPSA could, in the future, aid in identifying novel therapeutic targets for Alzheimer’s disease, cancer, autoimmune disorders, and infectious disease, including Ebola, influenza, Zika, and more.
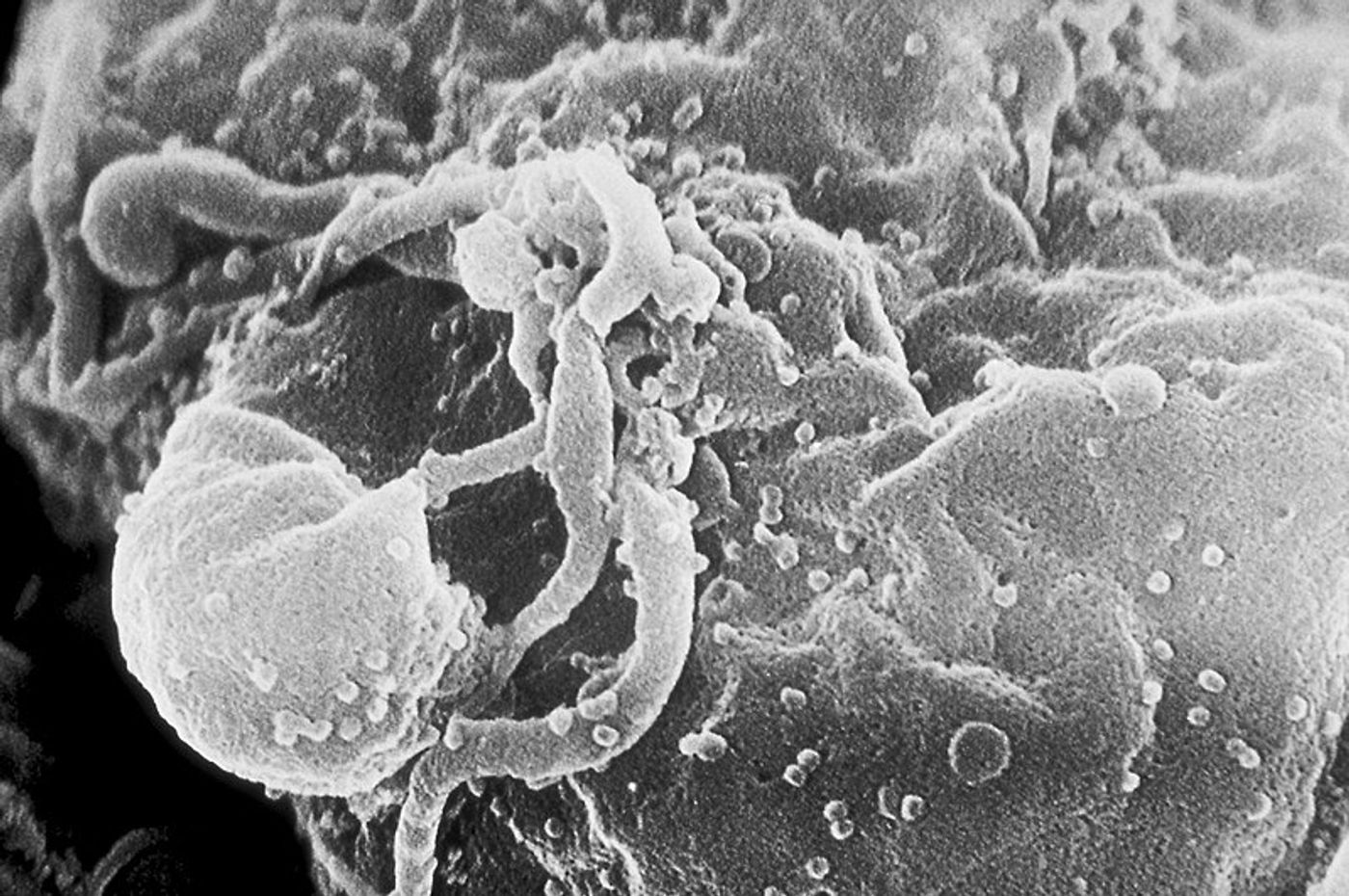
GAPSA helped scientists identify proteins in the human host that an HIV accessory protein called Vpu targets as a way to infect the cell and promote replication. Vpu destroys host proteins whose very function is to protect the cell from HIV infection. Vpu also works alongside another HIV accessory protein, Vif, which regulates viral infectivity.
"We selected Vpu as a test case because although some Vpu targets were known, we suspected there were more,” explained senior author Sumit Chanda, PhD. “Indeed, GAPSA was able to pinpoint several host proteins with anti-viral activity that had not been reported in connection with HIV."
In total, Chanda and his team screened 433 interferon-stimulated genes (ISGs), which activate in response to an infection. The screened each against Vpu to “create a more comprehensive list of HIV’s cellular targets,” explained co-author Lars Pache, PhD.
What can identifying Vpu targets with GAPSA do for the anti-HIV agenda? Researchers hope that they can translate their findings into the discovery of new drugs to block Vpu-directed protein destruction, enabling these proteins do their job and protect the body from HIV infection.
“In addition to providing critical knowledge of how cells work, the technology can be applied to identify protein degraders that specifically target disease-causing proteins, which can open new therapeutic opportunities for a multitude of diseases," Chanda explained.
Chanda, Pache, and others hope to continue their research by communicating with scientists in different fields to identify molecular circuits that regulate protein stability in the contexts of different diseases.
“We plan to use the the technology to comprehensively catalog pairs of all human proteins known to regulate degradation and their cellular targets,” Chanda said. “We anticipate that this compendium of activities will expand the therapeutic landscape for many diseases."
The present study was published in the journal Cell Reports.
Sources: Methods in Molecular Biology, Sanford-Burnham Prebys Medical Discovery Institute