Flesh-Eating Infection Turns Deadly When Two Microbes Are to Blame
Some bacterial infections get serious quickly and can be very difficult to treat, and scientists have found that these aren't always due to antibiotic-resistant microbes; they can happen because a person is infected with more than one microbial pathogen, and those pathogens can interact with one another. These so-called polymicrobial infections can be difficult to diagnose compared to illnesses caused by one microbe, or monomicrobial infections. Now a research team has used genetic tools to demonstrate that two strains of flesh-eating bacteria can be more dangerous than either is alone. The findings have been reported in the Proceedings of the National Academy of Sciences (PNAS).
"This research provides clear evidence that a very severe infection considered to be caused by a single species of a naturally occurring bacterium actually had two strains," said study co-author Rita Colwell, a Distinguished University Professor in the University of Maryland Institute for Advanced Computer Studies. "One of the strains produces a toxin that breaks down muscle tissue and allows the other strain to migrate into the blood system and infect the organs."
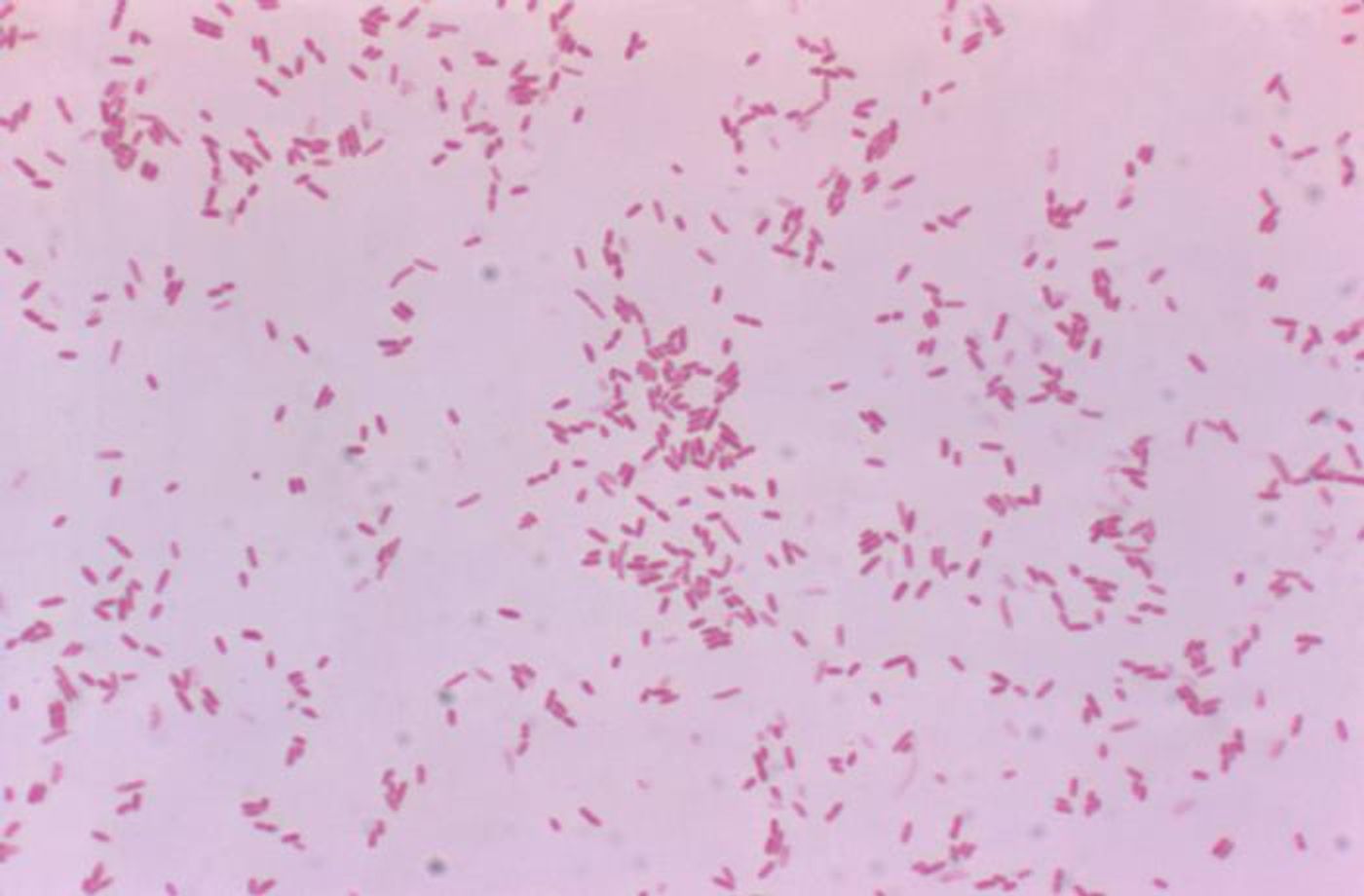
The scientists were able to analyze samples taken from a patient that had a flesh-eating disease called necrotizing fasciitis, which had been diagnosed as a monomicrobial infection caused by the bacterium Aeromonas hydrophila (according to the Centers for Disease Control and Prevention, other types of bacteria can also cause necrotizing fasciitis). The illness turned deadly serious and required a quadruple amputation to save the life of the patient.
In previous work, Colwell's team found that there were two strains of pathogens that are genetically distinct but can both cause the flesh-eating illness; the strains were dubbed necrotizing fasciitis 1 (NF1) and necrotizing fasciitis 2 (NF2). Neither strain results in a serious infection by itself. When they were combined, however, they became deadly.
In this work, after manipulating the genetics of the NF1 and NF2 microbes, the researchers could make one behave like the other. Mouse studies showed that the strains impacted each other, altering their ability to cause infection.
NF1 does not spread to the bloodstream on its own, and the immune system can get rid of it. NF2 can generate a toxin that can attack muscle tissue and enables the bacterium to spread to other places, including the bloodstream. When the pathogens get into the same host, the NF2 toxin still breaks muscle tissue down, but it also allows NF1 to migrate to the bloodstream and other organs, where it can be deadly. In these cases, NF2 doesn't move to other parts of the body.
"We're excited by this very elegant detective work," Colwell said. "We now have the ability through metagenomics to determine the individual infectious agents involved in polymicrobial infections. With these powerful new methods, we can determine how microbes work together, whether they're bacteria, viruses or parasites."
If polymicrobial strains that cause infection can be identified, it's far more likely that treatment can be successfully applied and improve patient outcomes. Missing one pathogen in a polymicrobial infection may cause chronic infections that are resistant to treatment. Colwell noted that a combination of therapeutics may be necessary to stop these infections. By using the genetic analysis developed in this research, clinicians may be able to identify and target polymicrobial infections.
"When we treat with a given antibiotic, we're clearing an organism out of the body," Colwell said. "But if there's another organism that's participating in the infection and that's also pathogenic, then any antibiotic treatment that doesn't also target that organism may just be clearing ground for it to grow like crazy."
Site: Phys.org via University of Maryland, PNAS