Antibiotic Resistance Genes Can Come From Random DNA Sequences
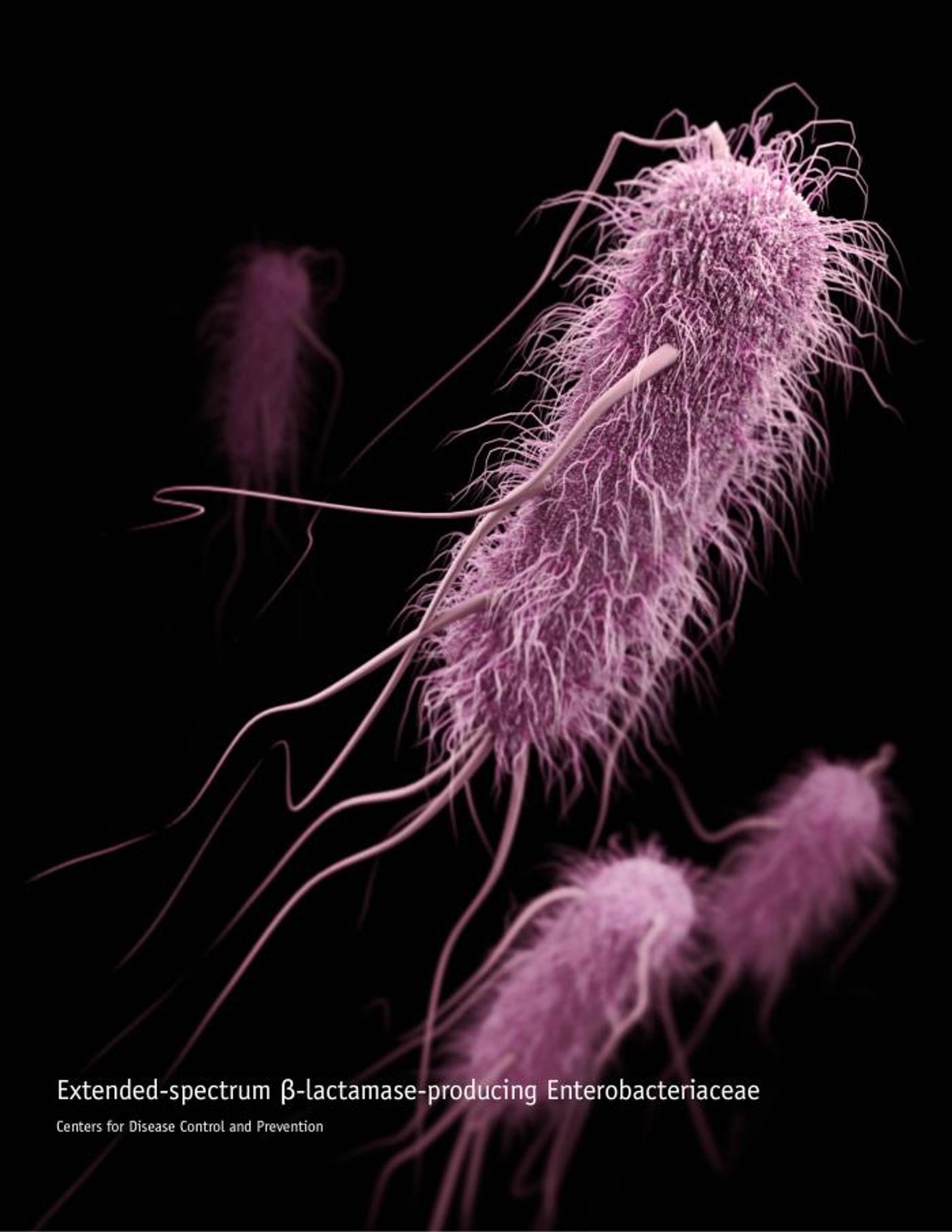
Bacterial infections that can’t be eliminated with standard drugs, caused by antibiotic-resistant bacteria, are considered to be a major and growing threat to public health. The pandemic has shown us how important it is to be able to contain infectious pathogens and treat the illnesses they can cause.
Researchers can often use genetic data from pathogens to develop new therapeutic approaches. Genes can confer antibiotic resistance to bacteria, and many of these genes sit on mobile pieces of DNA that can be easily shared with other bacteria, causing antibiotic resistance to spread. But now researchers have also found that antibiotic resistance genes can arise on their own out of totally random sequences of DNA. The findings have been reported in PLOS Genetics.
Genes that confer antibiotic resistance encode for proteins that work in one of a few ways: they might simply work to neutralize, or eliminate the antibiotic from the bacterial cell; they may alter the target of the antibody so it’s no longer able to stop the growth of the pathogen; or they could reduce the concentration of the antibiotic so it’s no longer effective. Scientists try to identify and characterize new antibiotic resistance genes so that we’re aware of when new resistant bacteria are being generated, and can figure out what they do. These genes can also combine in one pathogen to create so-called super-bacteria that are resistant to multiple types of drugs.
In this work, the researchers aimed to determine whether random DNA sequences could potentially create an antibiotic-resistant microbe. They designed almost one billion random sequences of DNA that were put on plasmids. These plasmids, which are mobile pieces of genetic material just like those chared by bacteria, were then added to a harmless version of the bacterium Escherichia coli, which is often used in research laboratories. Once inside the bacterial cells, the machinery of the cell could turn these sequences into peptides - strings of amino acids like those that might be used to make a protein.
The researchers found that most of these random sequences had no impact on the microbes at all. But, they found six peptides that did have an effect; they enabled the E. coli to resist the effects of an antibiotic called Colistin. That drug is used as a last resort treatment for serious bacterial infections. It works by destroying the membranes of bacterial cells. However, these six peptides were each able to raise the expression of genes that changed the bacterial membrane such that the antibiotic could no longer bind to it and destroy it.
"We have now shown in two different studies that random sequences of amino acids can give rise to new functions that are beneficial to the bacterium such as antibiotic resistance. This suggests that the evolution of new functions from random DNA sequences is not as unusual as previously thought," said senior study author Dan I. Andersson, Professor in Medical Bacteriology at Uppsala University.
"An important question that remains unanswered and requires further study is whether these new genes are naturally present in bacteria or can only be observed in laboratory experiments," said the first study author Michael Knopp, a post-doctoral researcher at the Department of Medical Biochemistry and Microbiology at Uppsala.
Sources: AAAS/Eurekalert! via Uppsala University, PLOS Genetics