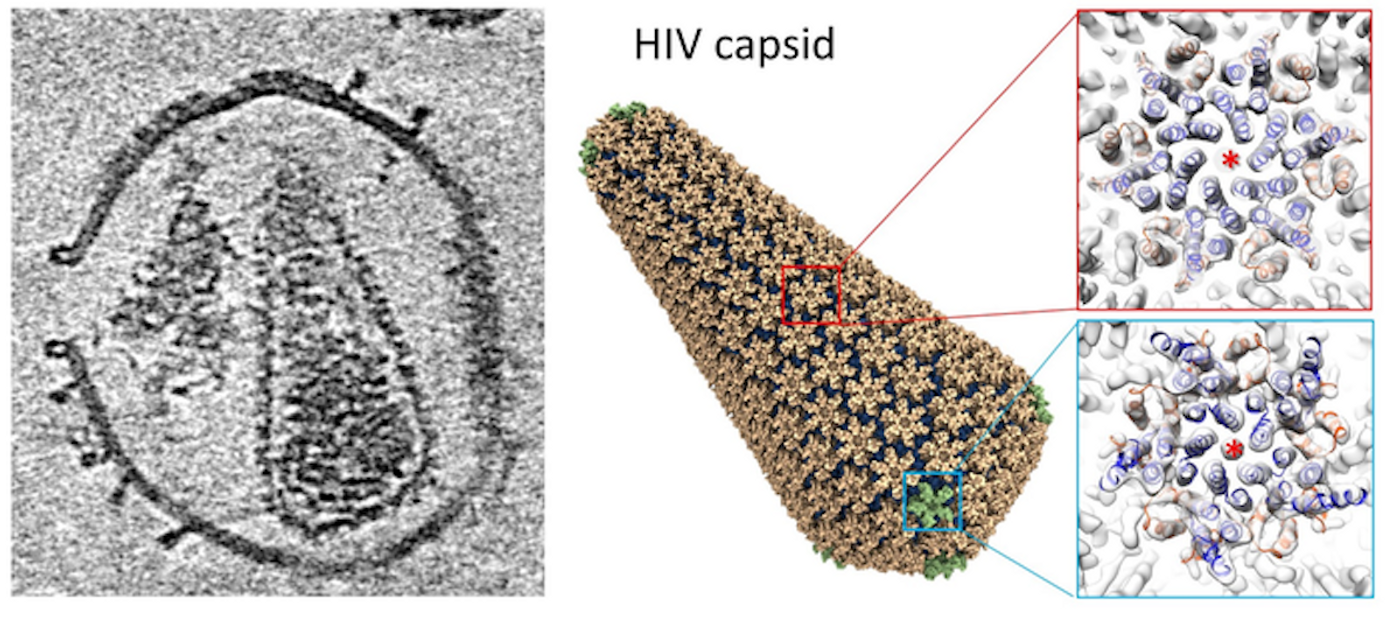
Researchers Solve the Structure of the HIV Capsid, May Be a New Drug Target
Though treatments are available, there is no cure or vaccine from HIV, which impacts about 38 million people worldwide. It's difficult to target the RNA genome of the HIV virus in part because it mutates so frequently, and as a retrovirus, it can insert itself into the genome of a host cell. The capsid shell that surrounds the viral genome of HIV could offer another targeting strategy, and researchers have now deciphered its complex structure. With cutting edge tools, scientists built a near-atomic level model of the HIV capsid. The work has been reported in Science Advances.
When HIV infects a host cell, the virus generates immature viral particles or virions with a dynamic molecule called Gag polyprotein, which can move through maturation and proteolytic stages, changing conformation from an immature sphere to a complete conical capsid. This capsid has a variety of important roles in HIV-1 replication; it can protect the viral genome from the host immune system, promote entry into a host cell's nucleus, control intracellular transport, and encourage reverse transcription to enter the host genome. These functions are influenced by various molecules and host cell proteins.
To elucidate the structure of proteins, one has to have a sample. That's a challenge for the HIV-1 capsid, which changes shape and is difficult to isolate intact in large quantities. Typical methods that are used to purify viral capsids aren't effective; they cause the HIV-1 capsid to dissociate. In this study, the researchers developed a new strategy that does not use detergent like the standard method.
“Instead of detergent extraction, we punctuate the membrane of HIV virus-like particles with a pore-forming toxin, which avoids the trauma associated with lysis of the virions and isolation of the cores, but also makes the capsid accessible to external cell factors and small molecules," said lead study author Dr. Tao Ni of the University of Oxford.
The researchers were them able to use electron tomography and subtomogram averaging to analyze the interactions between the HIV-1 capsid and two molecules, Cyclophilin A (CypA), and cofactor IP6 (inositol hexakisphosphate). This work revealed the capsid's structure by itself, as well as its complex with CypA and IP6. There is a double IP6 binding site on the mature capsid, and the work revealed more about how these molecules help control the stability of the capsid.
“In collaboration with Professor Juan Perilla’s group in the University of Delaware, using information derived from electron tomography, we also built an atomistic model of the whole HIV capsid which could serve as a blueprint for the development of capsid-targeting antivirals. The perforation of the enveloped virus membrane also provides a novel approach to study host-virus interaction for other viral systems," said study leader Professor Peijun Zhang, Director of the eBIC at Diamond Light Source and a Professor of Structural Biology at the University of Oxford.
Sources: Diamond Light Source, Science Advances