Antibiotic resistance is becoming a serious public health problem. Figuring out how antibiotic resistance genes disperse is crucial to understanding how to prevent their dissemination. Humans directly interact with the environmental microbiota, which contains a stockpile of ancient and diverse antibiotic resistance genes, known as a resistome. This direct interaction between humans and the resistome facilitates horizontal gene transfer (the transmitting of genes between organisms that is not parent-to-child) which disperses resistance even further.
Many studies on environmental microbiomes and resistomes have focused on industrial environments or clean, remote settings. In
a new study published in Nature, scientists have taken a look at the bacterial community present in two different impoverished parts of the world near large urban centers. Poverty-stricken areas tend to have limited access to clean water and unregulated access to antibiotics, both of which dramatically increase the likelihood of pathogen transmission.
The researchers wanted to learn more about antibiotic resistance genes and bacteria in these locales by characterizing the organisms present in hundreds of samples taken from the environments of the areas under investigation, as well as fecal samples from residents. Using genetic sequencing to ascertain the composition of their samples, they found that antibiotic resistance is highly correlated with the type of setting, whether it be urban or rural, and is not just distributed at random across habitats.
They also found that microbiomes are highly dependent on the lifestyle of the host. The human microbiome has a high degree of variation and diversity that is dependent on myriad factors such as age, diet, culture and health. Despite huge differences in geography, like being on different continents, there was a high degree of similarity between microbiomes of hosts with comparable lifestyles. They determined that chicken feces in the environment creates a potent avenue for introducing antibiotic resistance genes that are compatible with human microbiota into soil, one example of how a person’s livelihood has a direct impact on their microbiome.
Interestingly, resistance to antibiotics was not correlated with industrialization. The highest degree of antibiotic resistance genes was found in a cohort described as a peri urban (the transition zone from urban to rural) community from Peru. The study coordinators had no problem detecting antibiotics in sewage samples they had collected either. The popular and readily available antibiotics chloramphenicol, ciprofloxacin, tetracycline, trimethoprim, and sulfamethoxazole were regularly detected during testing throughout the sampling period, in addition to other antibiotics found often.
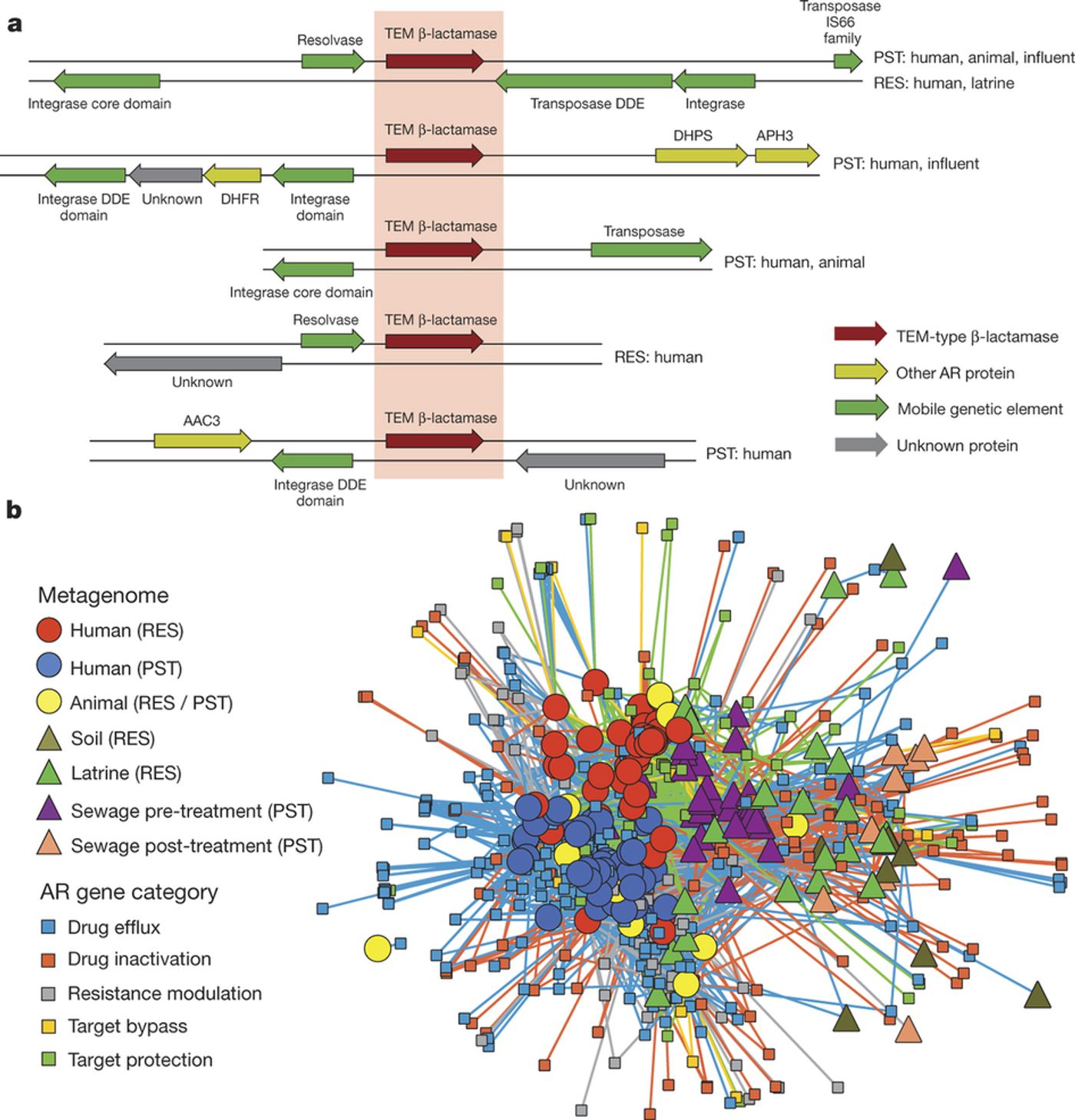
There was a small but nonetheless significant correlation between the proportion of antibiotic resistance genes with a mobile genetic element (MGE) or a multi-drug resistance gene (MDRC), supporting a role for MGEs and MDRCs in antibiotic resistance transfer across environments, in addition to increased accessibility to pathogens.
The authors suggest that future work could be critical for improving public health, especially by investigating the factors promoting or restricting antibiotic resistance exchange between environmental microbiota, humans, and pathogens. They highlight the treatment of waste as one such factor that merits further understanding.
Source:
Nature