Food poisoning sickens many people, and can lead to outbreaks of disease. Rapid detection of a common strain of pathogenic bacteria could prevent illness and stop the consumption of contaminated food more quickly. Three new technologies have been developed that could help public help officials reduce risk to the population.
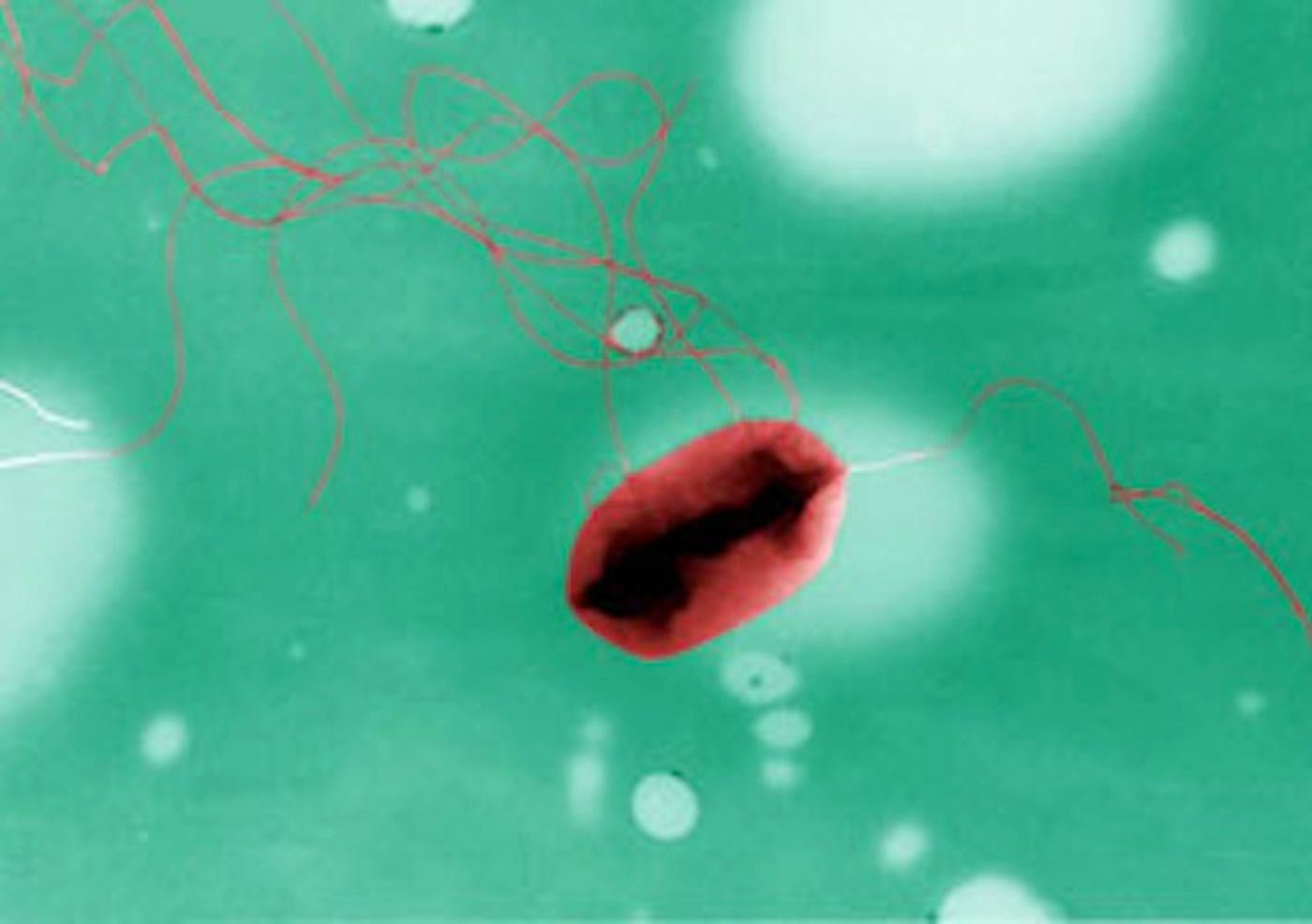
One bacterium these new tools are after is a specific strain of Escherichia coli, O157. While many strains of E. coli reside harmlessly in the gastrointestinal tract, and can even be beneficial, E. coli O157 has been linked to very serious infections in humans.
A new study performed at the University of Edinburgh's Roslin Institute used software to analyze the genetic information from samples that were taken from both people and animals, and learn the DNA signatures of E. coli samples that have resulted in infectious outbreaks in people. The software will then recognize the strains that sicken people when they are in animals.
The results have suggested to the researchers that fewer than 10 percent of E. coli O157 strains in cattle would likely cause illness in people. Steps could be taken to eliminate those strains in cattle as well, such as with vaccines.
"Our findings indicate that the most dangerous E. coli O157 strains may in fact be very rare in the cattle reservoir, which is reassuring. The study highlights the potential of machine learning approaches for identifying these strains early and prevent outbreaks of this infectious disease," explained Professor David Gally, senior author of
this work, which was published in the Proceedings of the National Academy of Sciences.
This approach is not limited to E. coli O157, the researchers say. A similar detailed analysis of microbes like salmonella could be undertaken to identify the strains that have potential to cause illness in humans.
Another study published in Infectious Diseases outlines a different tool; this one can be used to rapidly find varying levels of bacteria in a sample. Current methods that detect bacterial contamination take over 24 hours. Some, like magnetic resonance, detect small amounts of bacteria but aren’t effective at high bacterial concentrations. Fluorescent detection has the opposite problem.
A research team led by Tuhina Banerjee and Santimukul Santra at the Kansas Polymer Research Center, Pittsburg State University has decided to combine the two detection methods to improve it. They created a hybrid nanosensor to detect varying amounts of E. coli O157 in less than one hour. It was able to measure levels of E. coli in lake water.
In this case as well, the scientists say their device couold also be used (with modifications) to detect many pathogens besides E. coli.
One more device has been developed as well, described in the video above. This is a sensor created by investigators at the University of Quebec. The device,
reported in Optics Letters, is able to detect E. coli in water in less than 15 minutes.
"Pathogenic bacterial infection is one of the biggest causes of death, and a fast response time is much needed for timely detection and subsequent cure of bacterial infection," said Saurabh Mani Tripathi, a physicist at the Indian Institute of Technology Kanpur in India and one author of the work. "I'm excited by the very low time [our sensor needs] to accurately detect the presence of E. coli bacteria in water collected from environments at different temperatures."
Sources:
PNAS,
Science Daily,
Infectious Diseases,
Phys.org,
Optics Letters