Engineering Microbes for Potential use in Diagnostics
As we learn more about how our health is intimately connected to the microbes that live in our gastrointestinal tract, our microbiome, researchers want to take it into consideration when evaluating and developing treatment options. However, monitoring the microbiome is challenging. Reporting in Nature Biotechnology, scientists at the Wyss Institute of Biologically Inspired Engineering have created a bacterial sensor. Incredibly, it contains a gene circuit with sensor and reporter in a bacterial strain that it able to monitor gut inflammation, for six months when tested in mice. This work represents a step forward for diagnostics that can be used in patients.
Microbes have a natural ability to sense changes in their environment, and they can respond to those changes in a dynamic way. Normal microbes have already been introduced into the therapeutic realm; now researchers want to fully take advantage of those abilities, engineering and improving microbes so they can act as sensors, for example.
"We think about the gut as a black box where it is hard to see, but we can use bacteria to illuminate these dark places. There is great interest from patients and doctors that push us to build sensors for biomarkers of gut conditions like IBD and colon cancer," explained Pamela Silver, Ph.D, who led the work. "We believe that our work opens up enormous possibilities that can exploit the flexibility and modularity of our diagnostic tool and expand the use of engineered organisms to a wide variety of applications.”
Gene circuits, a set of genes that work together, must be introduced into a microbe in order for it to act as a sensor of biomarkers. When adding them into a bacterial genome, care must be taken to not disrupt any normal function of the microbe because it may cause death, and of course a sensor that will not function. Additionally, those alterations might not be fatal, but they often disrupt function enough that they are cleared from the body and not useful over long periods of time.
Related: Gene Expression Shown to be Influenced by the gut Microbiome
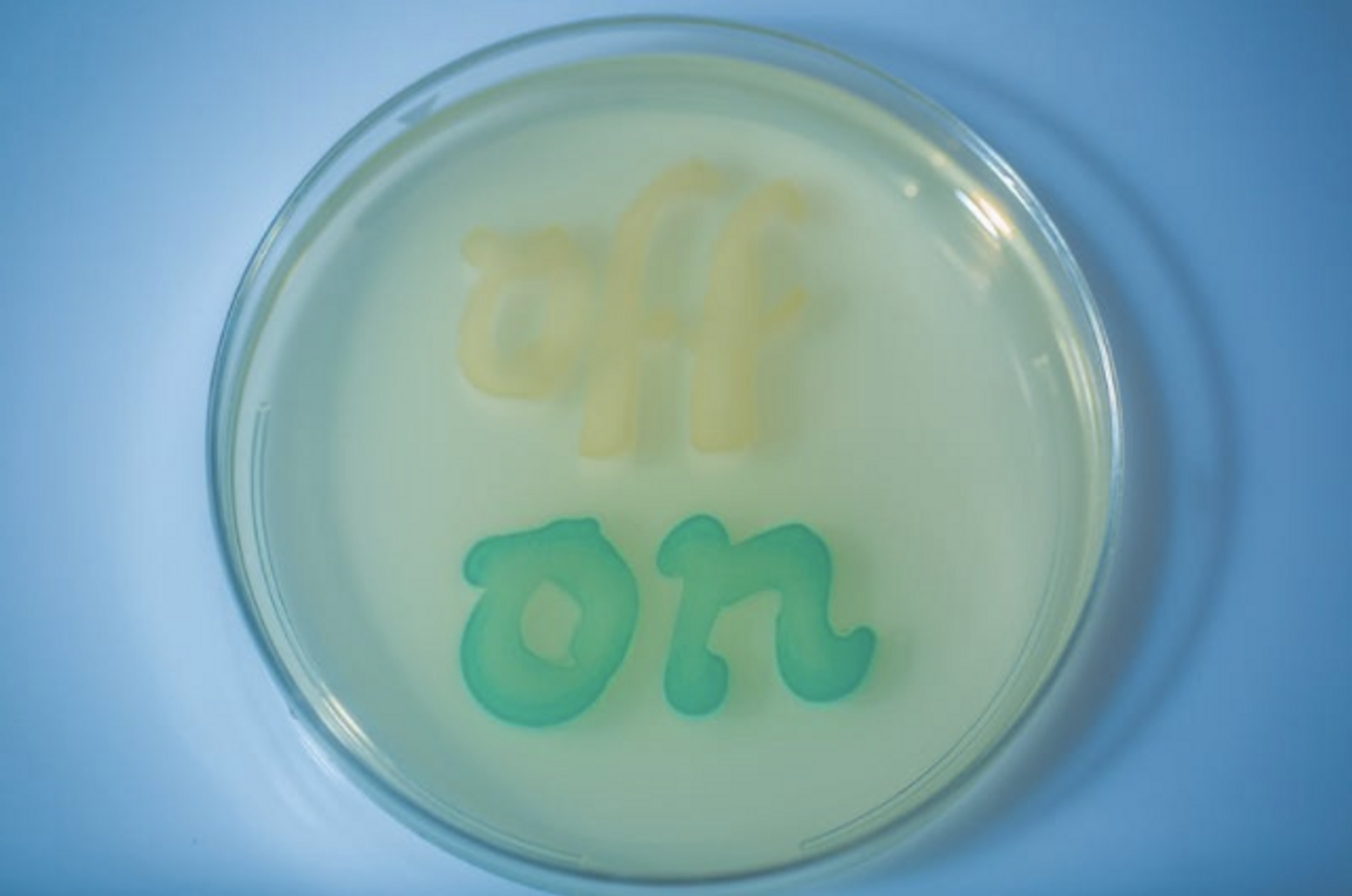
This new work aims to overcome those hurdles. The investigators constructed a memory module in the circuit that can detect a molecule, remaining responsive long after the stimulus has gone. The team engineered their bacteria, which is part of the mouse microbiome, to sense and retain levels of tetrathionate - a molecule made when the mouse intestine becomes inflamed.
Simply put, if tetrathionate is produced during inflammation and is sensed by the circuit, levels of one gene in the circuit decreases, a second gene is turned on, shutting off the expression of the first gene. A reporter is also present that makes the change easily detectable through a color change.
The sensor was tested for reliability, first in bacteria in culture, and the first author of the work, David Riglar, Ph.D., found that it could detect tetrathionate in mouse gut inflammation due to infection with a pathogenic bacterium, S. typhimurium. It continued to do its job for six months after it was administered. An easy analysis of fecal matter confirmed the memory of the synthetic circuit was on and its DNA was stable and unaltered.
"Our approach is to use the bacteria's sensing ability to monitor the environment in unhealthy tissue or organs. By adding gene circuits that retain memory, we envision giving humans probiotics that record disease progression by a simple and non-invasive fecal test," said Riglar.
"Pam's work demonstrates the power of synthetic biology for advancing medicine as it provides a way to rationally and rapidly design sophisticated sensors for virtually any molecule. If successful in humans, their technology would offer a much less expensive and more specific way to monitor gut function at home than sophisticated imaging instruments used today", said Donald Ingber, M.D., Ph.D., Founding Director of the Wyss Institute, a Professor of Vascular Biology at Harvard Medical School and at Boston Children's Hospital.
Learn more about the connection between the microbiome and inflammation from the following NIH lecture.
Sources: AAAS/Eurekalert! via Wyss Institute, Nature Communications