Understanding in vivo Metabolomics: C13 Isotope Studies
One key to understanding cancer metabolomics lies in the ability to accurately replicate the natural environment of the tumor in the lab. In vitro culturing of cancer cell lines is an easy way to initially understand the metabolic needs of specific cell lines and gene mutations. However, too often, drug targets identified in vitro, do not translate in vivo or in the clinic. The cellular and genetic heterogeneity and feedback mechanisms of the natural environment in vivo is missing in the in vitro setting. Therefore, it is important to also be able to accurately study real time metabolomics in the natural environment of the tumor.
C13 isotope analyses are a very useful tool for following the fates of nutrients inside the cell. In vitro experiments are easy to perform and can yield valuable insights. The push towards in vivo C13 labeling will allow researchers to confirm the cells preferred metabolic pathways and better understand druggable targets. The in vitro environment is lacking in the intricacies of a tumor environment. In vivo analyses often can reveal a switch in metabolic phenotypes due to limited availability of glucose or oxygen, things that are hard to accurately replicate in the in vitro environment.
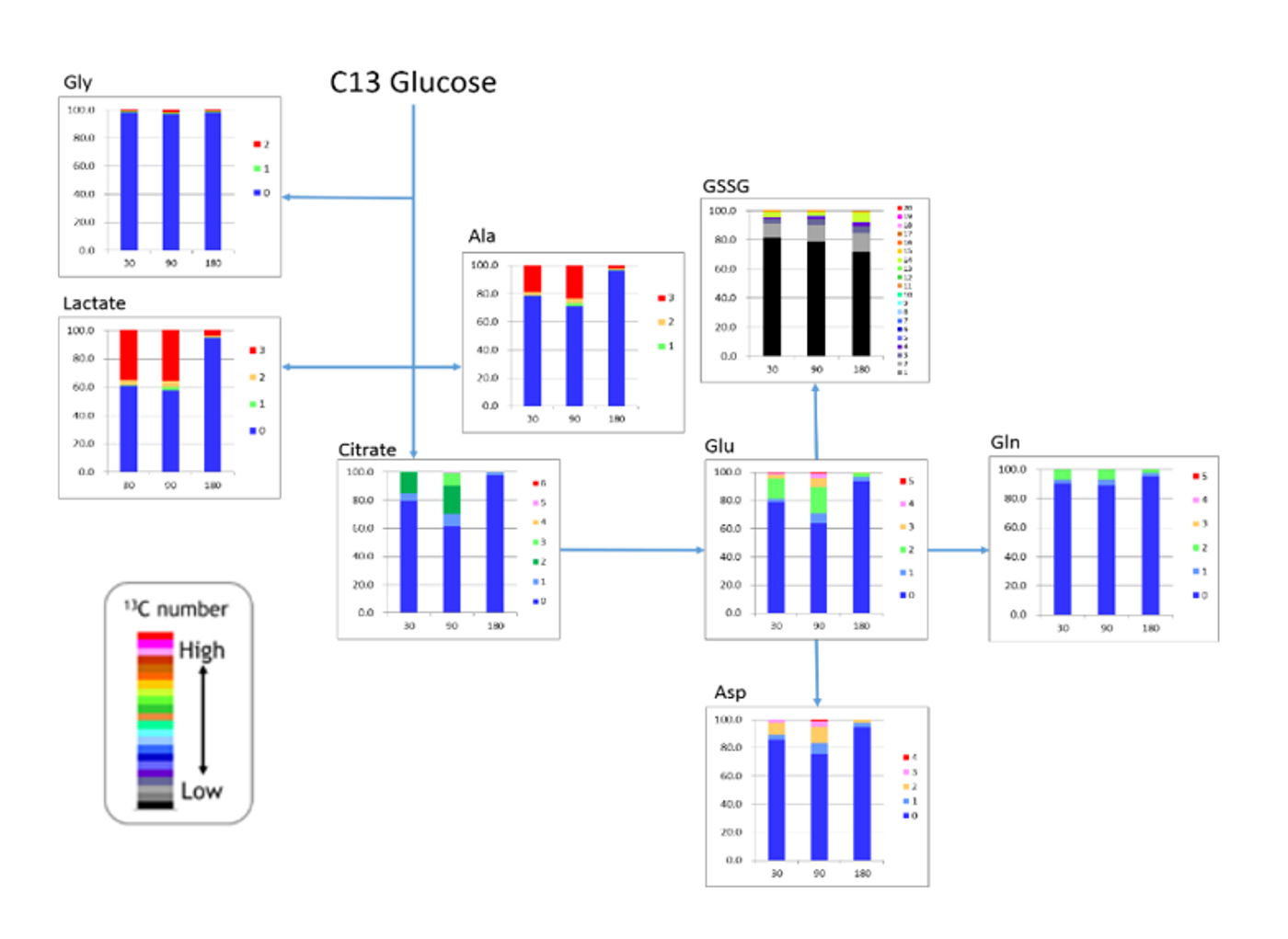
HMT is committed to providing our unique C13 analyses for cultured cells, as well as, human tumors in vivo. As a proof of concept (POC) experiment, we performed a time course analysis injecting a single dose of C13 glucose intraperitoneally into a mouse and analyzed subcutaneous tumors at 30, 90, and 180 minutes post injection for C13 incorporation into targeted metabolites.
In the figure, we observed rapid labeling of lactate that dropped off by 180 minutes. The number of labeled carbons is marked by the legend, illustrating in red, that all 3 lactate carbons were isotopically labeled with C13. Glycine is only labeled in a small portion (~2%), though it is well above the natural abundance. Glycine synthesis may be too slow to acquire a significant portion of the C13 glucose when it is injected as a single dose or these cells may not actively synthesize glycine, while both alanine and aspartate contained a significant level of label. We also see that the labeled carbons are entering the TCA as 2 carbon labeled acetyl CoA, thus resulting in 2 carbon labeled citrate (dark green). We also notice that the cells are actively synthesizing both glutamate, presumably for glutathione synthesis, as well as glutamine, again illustrated by the significant portion of 2 carbon label in these metabolites, as they will be produced from from the TCA.
Overall, we saw C13 incorporation into selected glycolytic intermediates, TCA cycle, amino acids, glutathione, and ATP. Future studies will focus on increasing the level of detection of incorporated C13, adding additional metabolites, and investigating the metabolic consequences of various anti-cancer agents. HMT also understands that every experiment is different, and we are working on developing additional labeling protocols based on the goals of the experiment.
As the push for better cancer therapeutics continues to rise, metabolomics will no doubt play a significant role. Metabolic therapies themselves are subject to varying responses, sometimes within the same cell line based only on whether or not the cells are tested in vitro or in vivo. It is imperative that we assess our systems both in vitro and in vivo for a better understanding of how these cells thrive, progress, and respond to cancer therapies.