Neutrons Enable Nanoscale-level View of a Live Cell Membrane
When researchers look at cells under high magnification, they usually have to look at cells that have been treated with special chemical reagents and are no longer alive. But a team of scientists at the Department of Energy's Oak Ridge National Laboratory has now examined a living cell membrane on a nanoscale. These new techniques using neutron scattering were reported in PLOS Biology and are briefly described in the following video. The work may provide a novel experimental method to study membrane biophysics and might be useful for the interrogation of other cell components as well. It has applications for research on drug and cell membrane interactions, biofuel and membrane, or antibiotic and membrane interactions.
This was a multidisciplinary undertaking led by biophysicist John Katsaras, chemist Bob Standaert and microbiologist James Elkins and was performed at the High Flux Isotope Reactor and Spallation Neutron Source of the lab, with the bacterium Bacillus subtilis. They showed that there are lipid clusters within the cell membrane, called rafts. It has been suggested that these lipid rafts contribute to signaling between cells and are vital to the functions of the cell. There had been uncertainty, however, about the very existence of lipid rafts that this research aims to settle.
"It became a debate," Katsaras said. "Some people believed they exist, while others believed they didn't. There was a lot of circumstantial evidence that could support either side."
Basically, there wasn’t a good way to see the lipid rafts to prove they were there. Neutron scattering analysis was used in place of light microscopy. Neutrons are not limited in the way that visible light is, and is able to give a view of the cell at the nanoscale. Not only that, it does not damage the cell as it is being observed. Because neutrons don’t carry a charge they are able to penetrate deeply into materials.
First there were some hurdles to be overcome if the scientists wanted this new tool to work. They needed to use neutrons that would scatter off of lipid molecules and not interact with other parts of the cell, and they had to find a way to tell the difference between various types of lipid molecules. The neutrons in deuterium, an isotope of hydrogen, turned out to be the answer. Unlike the nuclei of common hydrogen gas where there is a proton but no neutron, deuterium has a proton and a neutron in its nucleus. A cell doesn’t distinguish between these two gases, but by using neutron scattering, they look very different.
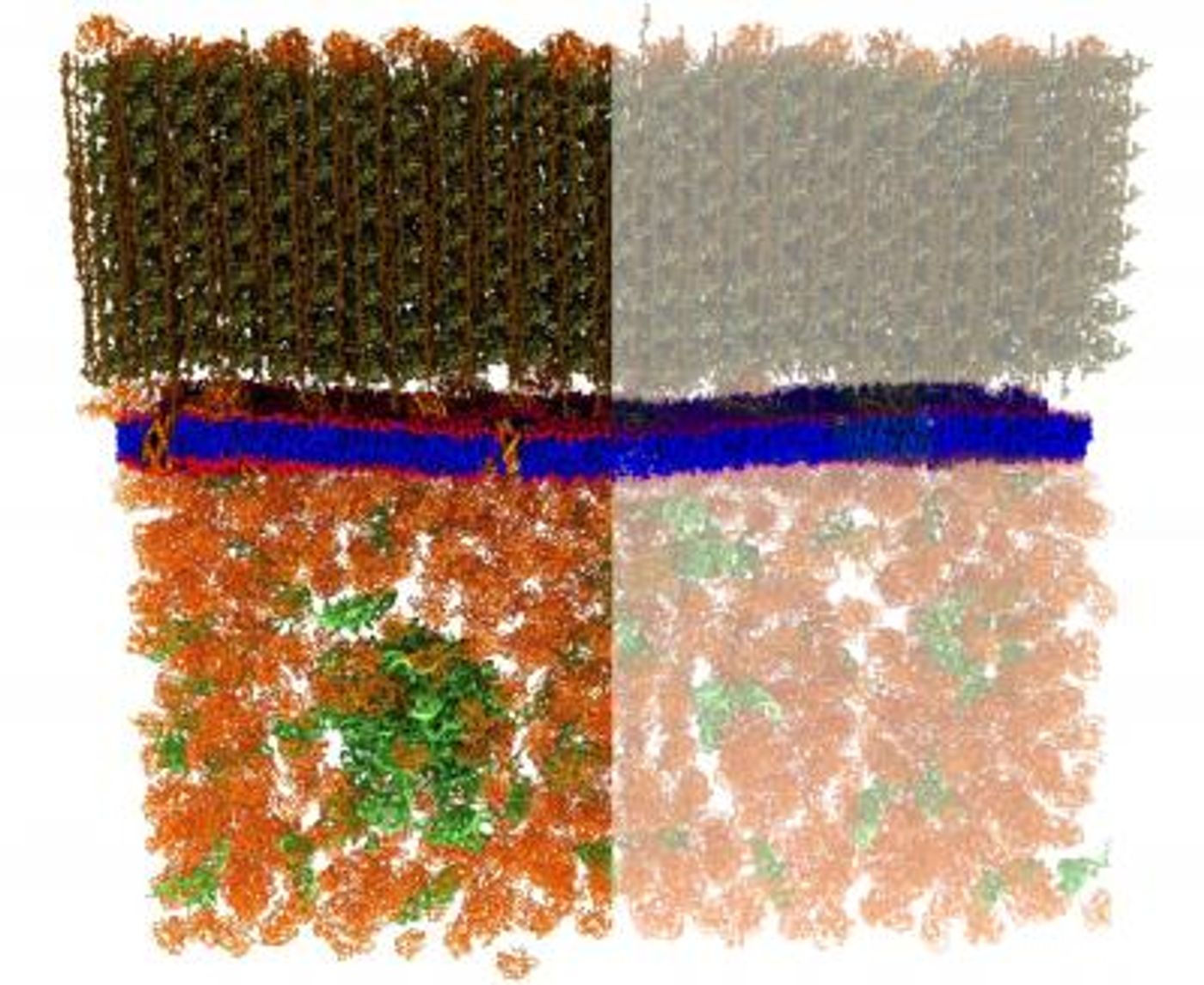
A special bacterial strain was engineered by the ORNL researchers - it contained enough deuterium that the structures of the cell would not be visible to neutrons. Then the bacteria were exposed to special lipid molecules that were made to contain fatty acids which had specific levels of deuterium and hydrogen. The bacteria were free to use those lipids to build membranes. If the lipids were randomly distributed in the membrane, the membrane would have a uniform appearance when exposed to neutrons, a grey image of even tone.
But, if lipids tended to group with other lipids that are like themselves, there would not be a uniform grey appearance, the image would have varying tones of lighter and darker grey areas. That turned out to be the case. The patches detected measured under 40 nanometers across, in a membrane roughly 2.4 nanometers thick. The ORNL researchers noted that the engineering they did to achieve internal contrast in live cells utilizing isotopes held potential for use in other research areas. It may open the method of targeted deuteration to other physical techniques such as nuclear magnetic resonance spectroscopy.
"The people who study these things tend to use particular types of probes," Katsaras noted. "They didn't use neutron scattering because it wasn't in the biologist's wheelhouse. Our novel experimental approach opens up new areas of research. For example, you could use the modified bacteria as a platform for investigating antibiotics, because a lot of these antibiotics really are talking to the membrane."
Sources: AAAS/Eurekalert! via ORNL, PLOS Biology

![Everything You Need To Know About NGS [eBook]](https://d3bkbkx82g74b8.cloudfront.net/eyJidWNrZXQiOiJsYWJyb290cy1pbWFnZXMiLCJrZXkiOiJjb250ZW50X2FydGljbGVfcHJvZmlsZV9pbWFnZV9mNTM1ZjIyYzA5MDE5ZmNmMWU5NmI0ZDc4NWU2MzdiZTZlN2I5ZDk5XzE4NDUuanBnIiwiZWRpdHMiOnsidG9Gb3JtYXQiOiJqcGciLCJyZXNpemUiOnsid2lkdGgiOjcwMCwiaGVpZ2h0IjozNTAsImZpdCI6ImNvdmVyIiwicG9zaXRpb24iOiJjZW50ZXIiLCJiYWNrZ3JvdW5kIjoiI2ZmZiJ9LCJmbGF0dGVuIjp7ImJhY2tncm91bmQiOiIjZmZmIn19fQ==)