Revolutionizing Our Approach to Antibiotic Resistance
Antibiotic resistance is already a major threat to public health; it's estimated that in 2019 alone, bacterial infections that evade the effects of antibiotics were directly responsible for the deaths of 1.27 million people or more, according to research reported in The Lancet. The problem is expected to get worse, as bacteria can easily evolve and share antibiotic resistance genes with one another. The various ways that antibiotics can be misused, such as when they are prescribed for things that won't be cured by antibiotics, can also encourage antibiotic resistance.
The search for new antibiotics is difficult, and while there has been some limited progress, researchers are always searching for new antibacterial drugs. Scientists have now found another approach.
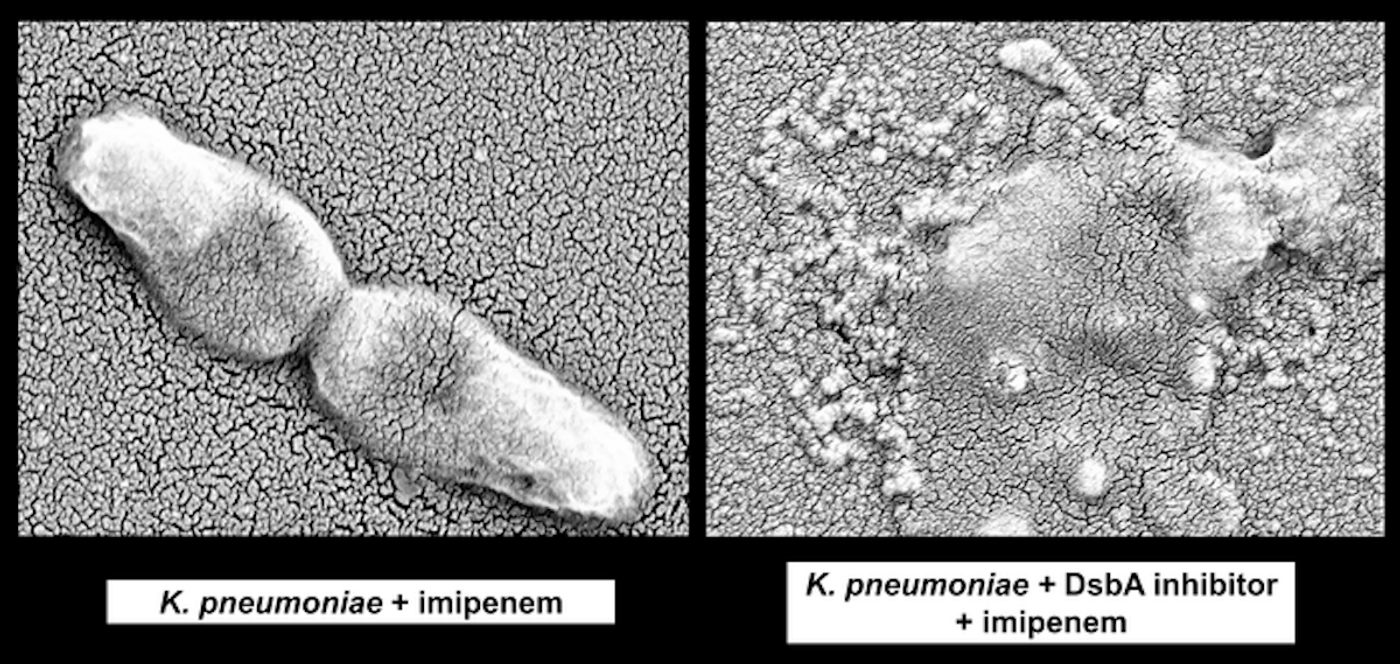
Reporting in eLife, investigators have suggested that bacteria can be made susceptible to antibiotics again by disrupting a protein that promotes antibiotic resistance in bacterial cells. By inhibiting this protein, called DsbA, it may be possible to make dangerous pathogens including resistant forms of E. coli, K. pneumoniae and P. aeruginosa, vulnerable again.
"It's a completely new way of thinking about targeting resistance," said study leader Despoina Mavridou an assistant professor of molecular biosciences at The University of Texas at Austin.
When bacteria express resistance genes, the proteins they encode for must be properly folded into a 3D shape for them to carry out their function; DsbA helps them fold into those forms. If DsbA is inhibited, crucial bonds in proteins don't form.
"Other approaches focus on inhibiting resistance proteins, but nobody had thought to try and prevent their formation in the first place," Mavridou said.
In this proof-of-principle study, the researchers disrupted DsbA with chemicals that would not be applied to humans. Now that their concept has been shown to be effective at making resistant bacteria vulnerable, they are looking for similar inhibitors that would be safe for people. If that approach is successful, the idea would be to combine the DsbA inhibitor with antibiotics that are already used, making them destroy bacterial infections once again. Since this method does not aim at a specific resistance gene or protein, but instead takes down the whole system, it could potentially eliminate the impact of several kinds of antibiotic resistance proteins.
This work has indicated that the modes of antibiotic resistance can be reversed in several pathogens by "targeting disulfide bond formation and protein folding," said study co-author Christopher Furniss of Imperial College London.
Sources: University of Texas at Austin, eLife