For Huge Numbers of Microbes, There is Still no Genetic Data
Technological advancements in computation and genetics have enabled researchers to sequence the genomes of vast numbers of organisms. With metagenomics techniques, researchers are able to sequence all of the DNA in a sample, then compare those genetic sequences to those that are known, and identify the organisms within the sample. Using tools like these, investigators have been able to catalogue many microbial species, and there have been efforts to learn more about microbes by analyzing environmental samples. But we share the world with vast numbers of microbes, and research has shown that we don't know much about most of them.
Evaluating the current state of our knowledge of microbial genetic diversity, researchers determined that for at least 42% of bacteria, there is no information about their genomes in current, public databases. This was a conservative estimate. The findings, which also propose a strategy for tackling the unknowns, have been reported in Science Advances.
"We took a deep dive into over 1.8 million bacterial and archaeal genomes to see how much of their diversity we've actually captured," said co-first study author Dongying Wu, of the Microbiome Data Science group at the Joint Genome Institute (JGI). "Turns out that despite all the genomes we've sequenced, we've only scratched the surface."
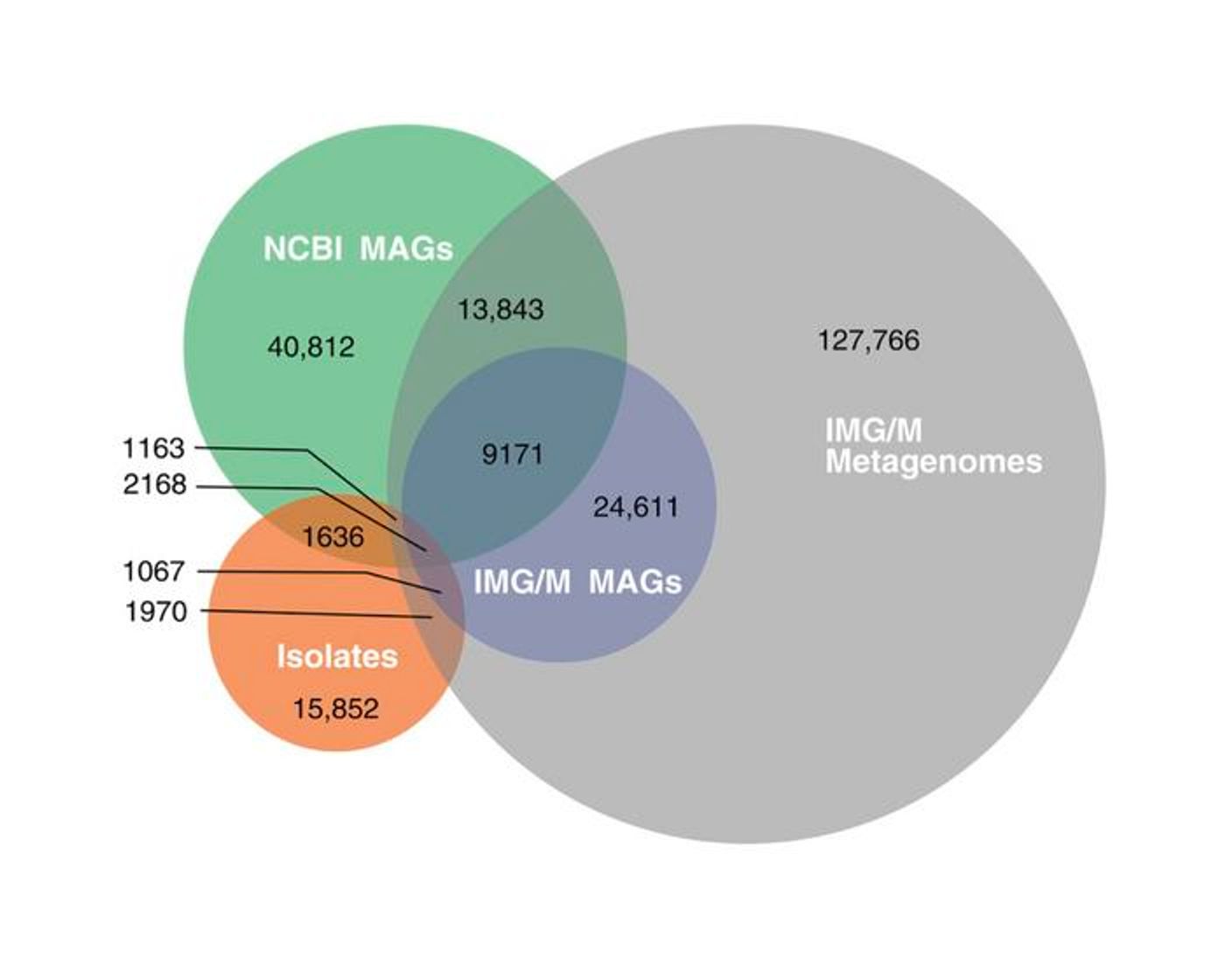
For this study, the researchers used phylogenetic diversity to represent the biodiversity of microbial species. They focused on five genes that can be found in nearly 2 million genomes, because they are universally conserved. The proteins are found in many sequences, including bacterial isolates and genomes that were collected in metagnomic studies, or metagenome-assembled genomes (MAGs).
Bacterial species that had been isolated and sequenced compose about 9.73% of the microbial diversity of current databases. About 49% of bacterial diversity has been captured in MAGs. For archaea, only about 6.55% of biodiversity has been gathered for isolates, while 57% is found in MAGs. So 36% of archaea are not represented at all.
Microbes are not only critically relevant to human health because of how they can affect us through the microbiome, or the microbial pathogens that can make us sick, they also influence global nutrient and chemical cycles. Soil microbes probably have a role in climate change, and microbes are also relevant to agriculture in many ways, from cultivation to consumption. Though metagenomic studies have broadly expanded what we know about microbial genomes, there is more work to be done.
"When it comes to isolate genome data, our real-world touchstones, we're just scratching the proverbial Petri dish," said co-first study author Rekha Seshadri, also of JGI. "It's a reminder of the urgency to cultivate new microbial species."
The metagenomic datasets that were analyzed in this work may also help scientists recover certain isolates, noted Seshadri. "We've drawn out the treasure map. Basically we can point specifically to environmental samples where people can go and reinvest time and effort in recovery."