Important fuels and chemicals have been made in genetically engineered strains of Escherichia coli by modifying this microbe’s fatty acid synthase (FAS), stimulating the bacterium to produce chemicals. E. coli has been altered to make chemicals used in the manufacture of detergents and lubricants as well as methyl ketones that have properties amenable to fuel production. However, up to this point, microbial production methods utilize tightly regulated enzymatic processes that can be inefficient for large scales.
Reporting in Metabolic Engineering, researchers with the U.S. Department of Energy's Joint BioEnergy Institute have developed an alternative FAS methodology that uses enzymes from other organisms to work with E. coli’s FAS, improving the microbe's capacity for making chemicals.
While fatty alcohols can be generated from plant, animal, and petroleum sources, using microbes for the synthesis of such chemicals is a much more sustainable method. Optimization of this system even more might lead to microbes with incredibly efficient conversion of biomass into fuels and other desired products.
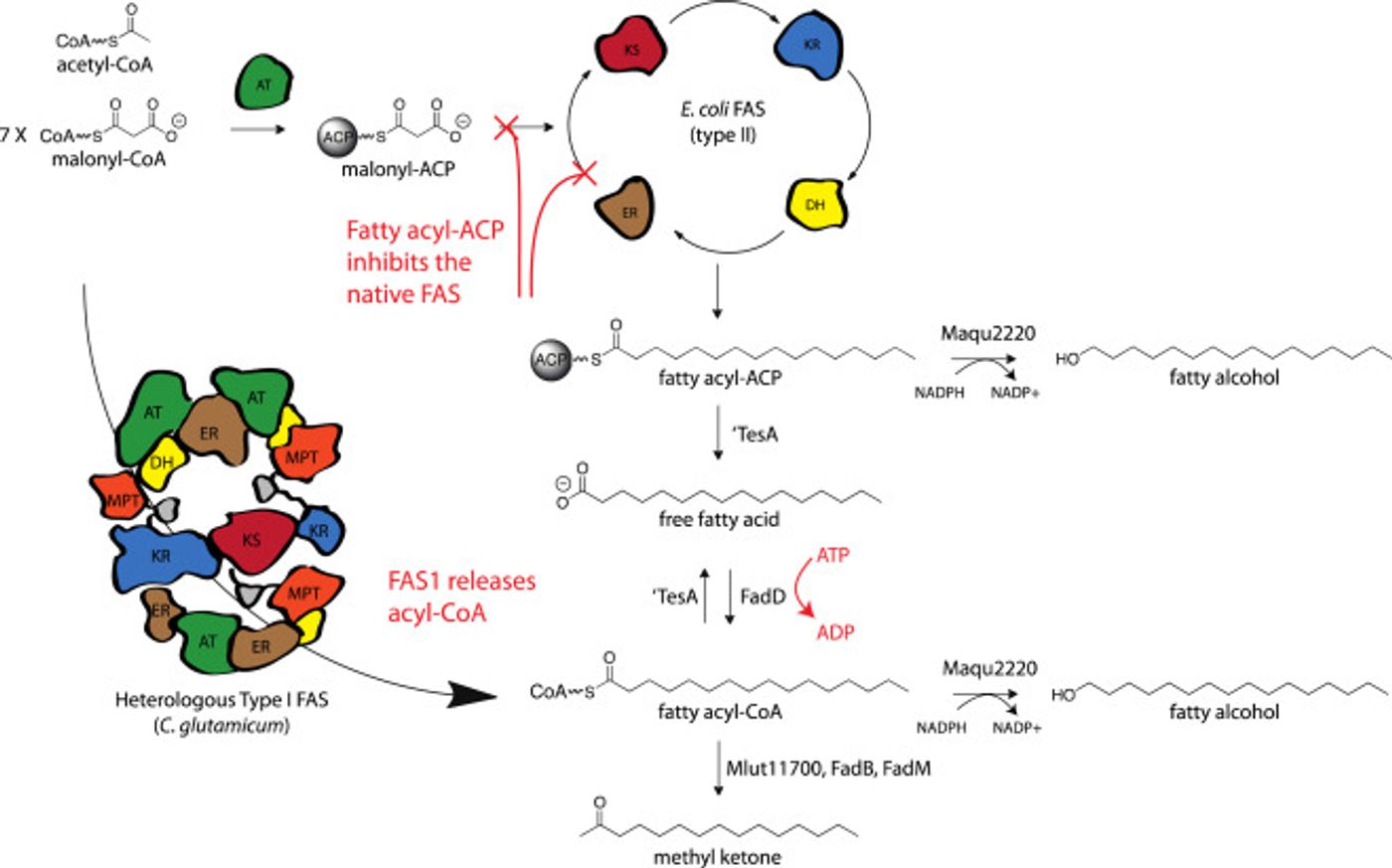
FAS is strictly regulated in bacteria as a vital part of metabolism. The BioEnergy Institute scientists got around that regulation by engineering several recombinant FAS enzymes that can function in parallel with the normal, native E. coli FAS. They determined that the engineered FAS that was the most active in E. coli was FAS1A from Corynebacterium glutamicum - a tiny, non-motile, Gram-positive soil bacterium. The investigators leveraged that enzyme to produce oleochemicals including methyl ketones and fatty alcohols.
C. glutamicum is well known as a workhorse in the fermentation industry. It’s been used for multi-million-ton scale production of glutamate and lysine for over 60 years. Relatively recently, research has focused on engineering it beyond simple amino acid manufacture.
It is believed that this work is the first example of in vivo function of recombinant, heterologous FAS in E. coli. The efficient use of FAS1 enzymes for biofuel and bioproduct production could potentially lead to the large scale conversion of biomass. In addition, the functional expression of such large enzyme complexes in E. coli will enable their study without needing to culture the native organisms.
The research team is currently investigating potential ways to make improvements to the system, and suggest several. Fatty alcohol production may be improved further by using E. coli strains that are engineered and selected for fatty alcohol tolerance. Another way might be to expand the repertoire of organisms from which FAS1 enzymes can be expressed, to vary genetic features, or to regulate of FAS1 expression dynamically, among other potential strategies the scientists are now exploring.
Sources:
Computational and Structural Biotechnology Journal,
Metabolic Engineering,
ScienceDaily via
Department of Energy, Office of Science


![[Guide] 7 Strategies to Boost Laboratory Collaboration](https://d3bkbkx82g74b8.cloudfront.net/eyJidWNrZXQiOiJsYWJyb290cy1pbWFnZXMiLCJrZXkiOiJjb250ZW50X2FydGljbGVfcHJvZmlsZV9pbWFnZV83YzBjZWIwM2Y5YzI4MmFlYzBhZDZhMTcyNTQ1ZGU3YmE4Y2MzMDYyXzUxNDkuanBnIiwiZWRpdHMiOnsidG9Gb3JtYXQiOiJqcGciLCJyZXNpemUiOnsid2lkdGgiOjcwMCwiaGVpZ2h0IjozNTAsImZpdCI6ImNvdmVyIiwicG9zaXRpb24iOiJjZW50ZXIiLCJiYWNrZ3JvdW5kIjoiI2ZmZiJ9LCJmbGF0dGVuIjp7ImJhY2tncm91bmQiOiIjZmZmIn19fQ==)